Abstract
Rare events have played an increasing role in molecular phylogenetics as potentially homoplasy-poor characters. In this contribution we analyze the phylogenetic information content from a combinatorial point of view by considering the binary relation on the set of taxa defined by the existence of a single event separating two taxa. We show that the graph-representation of this relation must be a tree. Moreover, we characterize completely the relationship between the tree of such relations and the underlying phylogenetic tree. With directed operations such as tandem-duplication-random-loss events in mind we demonstrate how non-symmetric information constrains the position of the root in the partially reconstructed phylogeny.






Similar content being viewed by others
References
Abascal F, Posada D, Zardoya R (2012) The evolution of the mitochondrial genetic code in arthropods revisited. Mitochondrial DNA 23:84–91
Ashkenazy H, Cohen O, Pupko T, Huchon D (2014) Indel reliability in indel-based phylogenetic inference. Genome Biol Evol 6:3199–3209
Bernt M, Merkle D, Rasch K, Fritzsch G, Perseke M, Bernhard D, Schlegel M, Stadler PF, Middendorf M (2007) CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 23:2957–2958
Boore JL (2006) The use of genome-level characters for phylogenetic reconstruction. Trends Ecol Evol 21:439–446
Boore JL, Brown WM (1998) Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev 8:668–674
Brandstdt A, Le VB, Rautenbach D (2010) Exact leaf powers. Theor Comput Sci 411:2968–2977
Bryant D, Steel M (1995) Extension operations on sets of leaf-labelled trees. Adv Appl Math 16:425–453
Calamoneri T, Sinaimeri B (2016) Pairwise compatibility graphs: a survey. SIAM Rev 58:445–460
Calamoneri T, Montefusco E, Petreschi R, Sinaimeri B (2013) Exploring pairwise compatibility graphs. Theor Comput Sci 468:23–36
Chaudhuri K, Chen K, Mihaescu R, Rao S (2006) On the tandem duplication-random loss model of genome rearrangement. In: Proceedings of the 17th annual ACM-SIAM symposium on discrete algorithms. ACM, pp 564–570
Deeds EJ, Hennessey H, Shakhnovich EI (2005) Prokaryotic phylogenies inferred from protein structural domains. Genome Res 15:393–402
Dekker MCH (1986). Reconstruction methods for derivation trees. Master’s thesis, Vrije Universiteit, Amsterdam, Netherlands
Donath A, Stadler PF (2014) Molecular morphology: higher order characters derivable from sequence information. In: Wägele JW, Bartolomaeus T (eds) Deep Metazoan phylogeny: the backbone of the tree of life. New insights from analyses of molecules, morphology, and theory of data analysis, chap. 25. de Gruyter, Berlin, pp 549–562
Durocher S, Mondal D, Rahman MS, (2013) On graphs that are not PCGs. In: Ghosh SK, Tokuyama T (eds) WALCOM: algorithms and computation: Proceedings of 7th international workshop, WALCOM 2013, Kharagpur, India, 14–16 February, 2013. Springer, Berlin Heidelberg, pp. 310–321
Dutilh BE, Snel B, Ettema TJ, Huynen MA (2008) Signature genes as a phylogenomic tool. Mol Biol Evol 25:1659–1667
Forst CV, Schulten K (2001) Phylogenetic analysis of metabolic pathways. J Mol Evol 52:471–489
Forst CV, Flamm C, Hofacker IL, Stadler PF (2006) Algebraic comparison of metabolic networks, phylogenetic inference, and metabolic innovation. BMC Bioinform. 7:67
Fritzsch G, Schlegel M, Stadler PF (2006) Alignments of mitochondrial genome arrangements: applications to metazoan phylogeny. J Theor Biol 240:511–520
Hartmann T, Chu AC, Middendorf M, Bernt M (2016) Combinatorics of tandem duplication random loss mutations on circular genomes. IEEEACM Trans Comput Biol Bioinform PP(99):1–1
Hellmuth M, Wieseke N (2015) On symbolic ultrametrics, cotree representations, and cograph edge decompositions and partitions. In: Xu D, Du D, Du D (eds) Computing and Combinatorics, vol. 9198 of Lecture Notes Computer Science. Springer International Publishing, pp 609–623
Hellmuth M, Wieseke N (2016) From sequence data including orthologs, paralogs, and xenologs to gene and species trees. In: Pontarotti P (ed) Evolutionary biology: convergent evolution, evolution of complex traits, concepts and methods. Springer, Cham, pp 373–392
Hellmuth M, Wieseke N (2017) On tree representations of relations and graphs: symbolic ultrametrics and cograph edge decompositions. J Comb Optim. https://doi.org/10.1007/s10878-017-0111-7
Hellmuth M, Hernandez-Rosales M, Huber KT, Moulton V, Stadler PF, Wieseke N (2013) Orthology relations, symbolic ultrametrics, and cographs. J Math Biol 66:399–420
Hellmuth M, Wieseke N, Lechner M, Lenhof HP, Middendorf M, Stadler PF (2015) Phylogenomics with paralogs. Proc Natl Acad Sci 112:2058–2063. https://doi.org/10.1073/pnas.1412770112
Hellmuth M, Stadler PF, Wieseke N (2016) The mathematics of xenology: di-cographs, symbolic ultrametrics, 2-structures and tree-representable systems of binary relations. J Math Biol. https://doi.org/10.1007/s00285-016-1084-3
Hennig W (1950) Grundzüge einer Theorie der Phylogenetischen Systematik. Deutscher Zentralverlag, Berlin
Hernandez-Rosales M, Hellmuth M, Wieseke N, Huber KT, Moulton V, Stadler PF (2012) From event-labeled gene trees to species trees. BMC Bioinform 13:S6
Krauss V, Thümmler C, Georgi F, Lehmann J, Stadler PF, Eisenhardt C (2008) Near intron positions are reliable phylogenetic markers: an application to Holometabolous Insects. Mol Biol Evol 25:821–830
Lafond M, El-Mabrouk N (2014) Orthology and paralogy constraints: satisfiability and consistency. BMC Genomics 15(S6):S12
Lavrov DV (2007) Key transitions in animal evolution: a mitochondrial DNA perspective. Integr Comp Biol 47:734–743
Mazurie A, Bonchev D, Schwikowski B, Buck GA (2008) Phylogenetic distances are encoded in networks of interacting pathways. Bioinformatics 24:2579–2585
Mehnaz S, Rahman MS (2013). Pairwise compatibility graphs revisited. In: 2013 International conference on informatics, electronics and vision (ICIEV), pp 1–6
Misof B, Fleck G (2003) Comparative analysis of mt LSU rRNA secondary structures of Odonates: structural variability and phylogenetic signal. Insect Mol Biol 12:535–47
Prohaska SJ, Fried C, Amemiya CT, Ruddle FH, Wagner GP, Stadler PF (2004) The shark HoxN cluster is homologous to the human HoxD cluster. J Mol Evol 58:212–217
Rogozin IB, Sverdlov AV, Babenko VN, Koonin EV (2005) Analysis of evolution of exon-intron structure of eukaryotic genes. Brief Bioinform 6:118–134
Rokas A, Holland PW (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15:454–459
Sankoff D, Leduc G, Antoine N, Paquin B, Lang BF, Cedergren R (1992) Gene order comparisons for phylogenetic inference: evolution of the mitochondrial genome. Proc Natl Acad Sci USA 89:6575–6579
Sempere LF, Cole CN, McPeek MA, Peterson KJ (2006) The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zoolog B Mol Dev Evol 306:575–588
Semple C, Steel M (2003) Phylogenetics, vol. 24 of Oxford lecture series in mathematics and its applications. Oxford University Press, Oxford
Shedlock AM, Okada N (2000) SINE insertions: powerful tools for molecular systematics. BioEssays 22:148–160
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
Stechmann A, Cavalier-Smith T (2003) The root of the eukaryote tree pinpointed. Curr Biol 13:R665–R666
Tang J, Wang LS (2005). Improving genome rearrangement phylogeny using sequence-style parsimony. In: Karypis G, Bourbakis NG, Tsai J (eds) Proceedings of the IEEE fifth symposium on bioinformatics and bioengineering (BIBE’05). IEEE Computer Society, Los Alamitos, CA, pp 137–144
Wagner G, Stadler PF (2003) Quasi-independence, homology and the unity of type: a topological theory of characters. J Theor Biol 220:505–527
Wang LS, Warnow T, Moret BM, Jansen RK, Raubeson LA (2006) Distance-based genome rearrangement phylogeny. J Mol Evol 63:473–483
Yang S, Doolittle RF, Bourne PE (2005) Phylogeny determined by protein domain content. Proc Natl Acad Sci USA 102:373–378
Yanhaona MN, Bayzid MS, Rahman MS (2010). Discovering pairwise compatibility graphs. In: Thai MT, Sahni S (eds) Computing and combinatorics: Proceedings of 16th annual international conference, COCOON 2010, Nha Trang, Vietnam, 19–21 July , 2010. Springer, Berlin Heidelberg, pp 399–408
Yanhaona MN, Hossain KSMT, Rahman MS (2008). Pairwise compatibility graphs. In: Nakano Si, Rahman MS, (eds) WALCOM: Algorithms and computation: proceedings second international workshop, WALCOM 2008, Dhaka, Bangladesh, 7–8 February, 2008. Springer, Berlin Heidelberg, pp 222–233
Acknowledgements
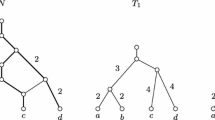
We thank the anonymous reviewers for the example in Fig. 2 and numerous valuable comments that helped us to streamline the presentation and to shorten the proofs. This work was funded by the German Research Foundation (DFG) (Proj. No. MI439/14-1 to PFS). YJL acknowledges support of National Natural Science Foundation of China (No. 11671258) and Postdoctoral Science Foundation of China (No. 2016M601576)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hellmuth, M., Hernandez-Rosales, M., Long, Y. et al. Inferring phylogenetic trees from the knowledge of rare evolutionary events. J. Math. Biol. 76, 1623–1653 (2018). https://doi.org/10.1007/s00285-017-1194-6
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-017-1194-6