Abstract
To discover bacteria with potential biocontrol activity against the rice sheath blight (RSB) pathogen, Rhizoctonia solani, cowdung (CD) associated bacteria were screened for antifungal activity a dual-culture method. Five potential biocontrol bacteria were identified to species-level based on their colony morphology, physiology, biochemical characteristics, utilization of carbon sources, micromorphology and 16S rRNA sequences. The ability of two selected strains to inhibit RSB was evaluated in vivo and in vitro. The median effective concentration (EC50) of crude extract from Streptomyces cochorusii strain NF0919 (NF0919) culture filtrate was 1.3 μg ml−1, lower than the EC50 of Jinggangmycin (a commercial antifungal agent widely used in China). At a concentration of 25.0 μg ml−1, the crude extract completely inhibited mycelial growth of R. solani. The field biocontrol efficacy after spraying 7 days in 2013 and in 2014 was 78.4 and 98.1% with crude extract from NF0919 culture filtrate and 71.1 and 94.2% with fresh cells of Bacillus amyloliquefaciens strain SB177 (SB177). Results from the 2-year field experiment suggested that the crude extract from NF0919 culture filtrate or fresh cells of SB177 provided better disease control than other fungicides (Jinggangmycin and/or Kresoxim-methyl). S. cochorusii strain NF0919 and B. amyloliquefaciens strain SB177 have good potential for field application and commercial use against the RSB pathogen.
Similar content being viewed by others
Introduction
Rice sheath blight (RSB), caused by Rhizoctonia solani kühn, is one of the main rice diseases worldwide [28, 35, 54]. Yield losses range from approximately 2.5–50% [46]. R. solani survives in soils as sclerotia, so it is very difficult to control effectively with chemical fungicides [12, 13, 48]. In the long run, chemical fungicides spaying also will have adverse environmental impact. Efficient and environmentally benign agents are urgently required to control RSB.
Breeding disease-resistant rice cultivars is believed to be one of the most promising approaches to control the disease. However, no rice cultivar has been found completely resistant to the soilborne fungus so far [5, 69]. Biocontrol of RSB has been reported and well documented. Biological control of sheath blight can be achieved by using antagonistic Pseudomonas spp. [38–40, 47, 56], Bacillus spp. [9, 10, 30, 42, 59, 60, 63, 67, 68], Trichoderma spp. [49, 53, 57], and antifungal metabolites produced by Streptomyces spp. [2, 21, 31, 43, 44, 50, 62, 65].
Despite the abundant biocontrol reports, few commercial formulations of biocontrol agents have been used successfully to control the RSB. Environmental constraints (ultraviolet light, different types soil, soil or air humidity, temperature) and disadvantageous formulation characteristics (short shelf-life, uncertain content, slow activity) have limited the use of biocontrol formulations with fresh cells (or spores), especially for field applications. To our knowledge, the most successful antifungal antibiotics for control of RSB in China and Japan, respectively, are Jinggangmycin from Streptomyces hygroscopicus var. jinggangensis 5008 [50] and Validamycin from S. hygroscopicus var. limoneus [21]. Jinggangmycin has been used for 35 years in China and has become the sole antibiotic used to control sheath blight. Some reports have noted a decline in the field control efficiency of Jinggangmycin and some tolerance in isolates of R. solani [19, 66].
Our investigation was carried out to discover more antifungal biocontrol resources. The purpose of this study was to (a) screen and identify the potential biocontrol ability of bacteria cultivated from CD, (b) find new bacteria able to prevent RSB, and (c) evaluate the potential biocontrol agents (fresh spore suspensions of Bacillus spp. and crude extracts of metabolites from Streptomyces spp.) in vivo and in vitro conditions.
Materials and methods
Isolation and selection of antagonistic bacteria from cowdung
Fresh CD was sampled from lactating cows and brought in sterile plastic bags to the Microbiology Laboratory of Zhenjiang Institute of Agricultural Science (Zhenjiang, Jiangsu Province of east China). Ten-gram samples of CD were transferred separately to sterile Erlenmeyer flasks containing 90 ml sterile PBS solution (PBS; containing 1.2 g Na2HPO4, 0.18 g NaH2PO4, 8.5 g NaCl, made up to 1 l with distilled water, pH 7.2). The samples were shaken for 30 min at room temperature. The suspensions were filtered through doubled, sterile gauze prior to isolation of bacteria. The filtered solutions were serially diluted to 10−1–10−5 with sterile PBS, and 0.1 ml of each dilution was spread on R2A medium (R2A; containing 0.75 g peptone, 0.50 g dextrose, 0.30 g dipotassium phosphate, 0.50 g starch, 0.50 g yeast extract, 0.30 g sodium pyruvate, 0.25 g tryptone, 0.024 g magnesium sulfate, and 15.00 g bacteriological agar, made up to 1 l with distilled water and adjusted to pH 7.2) and on Gause’s synthetic media to screen bacteria and actinomycetes, respectively. Three replicates (Petri plates) were prepared for each dilution and each incubation temperature. The plates were incubated for 3–5 days at 20 °C, 25 °C, or 30 °C to assay for a wide spectrum of bacteria. The antagonism of all bacterial isolates was checked with respect to their ability to suppress the growth of R. solani ZJ-1 (provided by Jiangsu Academy of Agricultural Science), inoculated on Waksman agar (WA; containing 5 g proteose-peptone, 10 g glucose, 3 g meat extract, 5 g NaCl, 20 g agar, made up to 1 l with distilled water and adjusted to pH 6.8). After 5 days of incubation at 25 °C, zones of inhibition were measured as described [4]. Eleven strains of bacteria (JR08, Mg116, RM09, SB177, NF0912, NF0919, NF0931, NF0939, NF0945, NF0974, NF0998) were finally selected based on their antagonistic activity toward fungi (measurement of inhibition zones). All the antifungal isolates were purified twice and then stored at −80 °C in nutrient broth containing 15% glycerol for further use.
Elucidation of antifungal spectrum of the selected antagonists
A dual-culture plate method was used to detect the antifungal spectrum of the selected antagonists. The antifungal activity of all bacterial isolates was checked by examining their ability to suppress the growth of the following phytopathogens from the main crops in Jiangsu province: Penicillium expansum JR21 (causes apple blue mold), Gloeosporium fructigenum XX43 (causes anthracnose of grape), Rhizopus nigricans GS84 (causes strawberry black mold rot), Gibberella zeae NA49 (causes Fusarium head blight on wheat), Fusarum oxysporium SW2 (causes strawberry wilt), Botryosppuaeria berengeriana YN19 (causes ring rot of apple), Fusarium oxysporum HB16 (causes cotton fusarium wilt), Magnaporthe grisea 04-025-4 (causes rice blast), Glomerella cingulata DS11 (causes apple anthracnose), F. oxysporum DT04 (causes watermelon wilt), Verticillium dahliae XJ37 (causes cotton verticillium wilt), Botrytis cinerea BC08 (causes grape gray mold), R. solani ZJ-1 (causes RSB), and Sclerotinia sclerotiorum JY20 (causes white mold of canola). The phytopathogens were provided by Jiangsu Academy of Agricultural Science. All the phytopathogens were inoculated in separate plates on WA. After 5 days of incubation at 25 °C, zones of inhibition were measured as described [4].
Bacterial characterization based on production of hydrolytic enzymes and secondary metabolites
Production of cell wall degrading enzymes and antifungal secondary metabolites are common mechanisms that bacteria use to inhibit fungal growth. To better characterize the antifungal isolates, their potential production of hydrolytic enzymes and secondary metabolites was studied. Previously described methods were used to assess chitinolytic activity in minimal medium [11], siderophore production [51], amylase activity [14], cellulase activity [16], phosphorus dissolution [25], and nitrogen fixation [22]. Protease activity indicated by casein degradation was determined from distinct zones of clearing in skim milk agar (50 ml sterilized skimmed milk mixed at 55 °C with 50 ml of 1/5 Waksman medium and 2% agar) after 3 days’ incubation at 30 °C.
Identification of the selected isolates
Five strains of the selected antifungal bacteria were identified taxonomically based on colony morphology, physiological and biochemical characteristics as described in Bergey’s Manual of Systematic Bacteriology [15], utilization of carbon sources [17], micromorphology [7], and 16S rRNA gene sequences [64].
Bioassay of antifungal activity of S. cochorusii strain NF0919
For detection of bioactive metabolites, a loopful of cultured S. cochorusii strain NF0919 was inoculated in Gause’s synthetic broth and incubated on a rotary shaker (200 rpm) at 30 °C as a seed culture. After 48 h of incubation, the seed culture (10% V/V) was transferred to a 30-l fermentor (Zhenjiang Dongfan GUS-30), containing 18 l optimized fermentation medium (potato starch 8.5%, cottonseed protein 2.2%, CaCO3 0.1%, K2HPO4 0.05%, and MgSO4 0.05%, W/V). The culture medium was agitated at 360 rpm, and the aeration flow was maintained at 3.2 air-volume/liquid-volume/minute (VVM). The fermentation was carried out for 5 days at 30 °C. The fermented culture was centrifuged at 10,000 rpm for 10 min, and the supernatant was filtered through a 0.25 μm filter. To obtain the crude extract, the culture filtrate was extracted twice with ethyl acetate and concentrated to dryness under a vacuum at 35 °C.
The crude extract (dissolved in a minimal volume of dimethyl sulfoxide) and 60% Jinggangmycin (as a positive control) were diluted to 0.78125, 1.5625, 3.125, 6.25, 12.5, 25.0, and 50.0 mg l−1 and mixed with molten WA medium, respectively. Ten milliliters of each treatment mixture was poured into sterile Petri plates (7 cm diameter). Agar plugs (5 mm diameter) taken from a 3-day-old WA culture of R. solani were placed on the WA plates as inocula. All the plates were incubated in dark at 25 °C. A culture of R. solani on WA without an inhibitor was used as negative control. Triplicate plates were maintained for each treatment. Mycelial growth inhibition of R. solani was assessed as percent mycelial growth inhibition relative to the negative control after 60 h incubation.
Field trials of RSB control
A field experiment was carried out at the Zhenjiang Institute of Agriculture Science of Jiangsu province (eastern China) to evaluate the efficacy of a fresh cell formulation of B. amyloliquefaciens strain SB177 and a crude extract of culture filtrate from S. cochorusii strain NF0919 for the suppression of RSB. Five treatments were tested: (1) 5% (W/V) Jinggangmycin (generously provided by TongLu Biochemical Company, Zhejiang, China), 375.0 g a.i. ha−1 (500.0 mg a.i. l−1); (2) 50% Kresoxim-methyl (a chemical fungicide), 225.0 g a.i. ha−1 (300.0 mg a.i. l−1); (3) fresh cells of SB177 (1.0–1.2 × 109 CFU ml−1), 7.5 l ha−1 (1.0–1.2 × 107 CFU ml−1); (4) crude extract of strain NF0919 culture filtrate, 125.0 g a.i. ha−1 (167.0 mg a.i. l−1); and (5) water alone, 750.0 l ha−1 (untreated control). A rice field known to be naturally infested with R. solani (located in the Zhenjiang Institute of Agricultural Science test field) was divided into 15 plots; each plot was 20.0 m2. The planted rice cultivar was Nangeng 44, which is highly sensitive to R. solani. Each treatment included three replicates arranged in a randomized complete block design. No other fungicides were applied to the experimental plots. All other treatments, such as herbicides and fertilizers, were used in accordance with standard farm practices.
An inoculum of experimental bacteria (SB177) was prepared by growth in LB medium (LB; containing 10 g tryptone, 5 g yeast extract, 10 g NaCl, made up to 1 l with distilled water and adjusted to pH 7.5) for 24 h at 30 °C, 200 rpm, and adjusted to give a final cell concentration of 1.0–1.2 × 109 CFU ml−1. For field application, the suspension in LB was diluted with sterile-distilled water to 1.0–1.2 × 107 CFU ml−1. A crude extract of culture filtrate from NF0919 was produced as described above. The five treatments were sprayed with 750.0 l ha−1 at the rice booting stage. This field experiment was performed in 2013 and repeated in 2014.
Based on the percentage of stems with disease symptoms among 200 randomly selected rice plants, disease incidence of RSB was determined after treatment 7, 14, and 21 days, respectively. Severity of RSB was rated on a 0–9 scale using the Standard Evaluation System for rice developed by the International Rice Research Institute, Manila, Philippines [20].
Disease severity and control efficacy were calculated as follows:
-
Disease severity = [∑(the number of diseased plants in each scale × representative value of each scale)/(total number of plants investigated × the value of the highest scale)] × 100.
-
Control efficacy = [(increase in disease severity in the control − increase in disease severity in the treatment)/increase in disease severity in the control] × 100.
-
Increase in disease severity = (treatment disease severity − basic disease severity) × 100. Basic disease severity refers to the disease severity before treatments.
-
Data were subjected to one-way analysis of variance and compared using Duncan’s multiple range test (DMRT) at P < 0.01.
Nucleotide sequence accession numbers
The 16S rRNA gene sequences from five isolates were deposited in GenBank with accession numbers HM989896, HM989897, HM989898, HM989899, and HM989900.
Results
Screening of antagonistic bacteria against R. solani
The count of culturable bacteria in the fresh cowdung ranged from 6.6 × 105 to 1.4 × 106 CFU (cells g−1 fw). In total, 342 bacteria isolates were obtained and checked for antagonism against R. solani in an in vitro dual-culture plate assay. Forty-two (12.3%) of the isolates were antagonistic to R. solani. Of these antagonists, eleven isolates (NF0919, NF0912, NF0931, NF0939, NF0945, NF0974, NF0998, JR08, Mg116, RM09, and SB177) were selected for further studies based on the diameter of the inhibition zone (≥5 mm).
Elucidation of antifungal spectrum of eleven antagonists
Eleven antagonists (Table 1) were tested for inhibition of the 14 main phytopathogens of field crops and orchards in the Jiangsu province of China. Eight isolates (NF0912, NF0919, NF0939, NF0945, NF0998, Mg116, RM09, and SB177) were antagonistic to all 14 of the phytopathogens, producing distinct, wide zones (ranging from 1 to 20 mm) of inhibition of pathogen mycelium. Five of the antagonists (NF0919, JR08, Mg116, RM09, and SB177) showed the most effective inhibition of R. solani, with inhibition zones more than 15 mm.
Characterization of bacterial production of hydrolytic enzymes and secondary metabolites
Among the 11 selected antagonists (Table 2), no isolates expressed chitinolytic activity, inorganic phosphate dissolving activity, or organic phosphate solubilizing activity. All 11 isolates expressed proteolytic activity and amylolytic activity. Only three antagonists (SB177, RM09, and Mg116) showed siderophore activity and nitrogen fixing activity, and all seven selected actinomycetes (NF0912, NF0919, NF0931, NF0939, NF0945, NF0974, and NF0998) expressed cellulolytic activity.
Identification of five selected strains
Based on the antifungal activity and enzyme activity, five bacteria (JR08, Mg116, RM09, SB177, and NF0919) were chosen for taxonomic identification, and for the subsequent in vitro and field assays. Three strains (Mg116, RM09, and SB177) were identified as strains of Bacillus amyloliquefaciens according to the method described in Bergey’s Manual of Systematic Bacteriology [15] on the basis of morphological, physiological, and biochemical properties (Table 3), and on their 16S rRNA gene sequence (Table 4). Strain JR08 was identified as a Bacillus cereus strain by the same method. Strain NF0919 was identified as a strain of Streptomyces cochorusii on the basis of its culture characteristics (Table 5), physiology, biochemical characteristics, microscopic characteristic of sporophores (Table 6), and 16S rRNA sequence (Table 4).
Toxicity test of crude extract in vitro
The crude extract showed high toxicities against mycelia of R. solani. Mycelial growth of R. solani was completely inhibited by the crude extract at a concentration of 25.0 μg ml−1. In comparison, mycelial growth inhibition by Jinggangmycin (25.0 a.i. μg ml−1) was 62.5%. The EC50 was lower with crude extract than with Jinggangmycin, revealing that the inhibitory activity of the crude extract was stronger than the positive control (Table 7).
Biological control of RSB with antifungal bacteria in the field
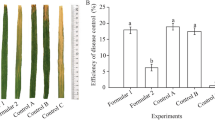
Rice plants were treated with foliar spray at the booting stage. In the 2013 field experiment, the percentage increase in disease severity in control plots at 7, 14, and 21 days after treatment was 15.7, 38.3, and 44.1%, respectively. The efficacy of biocontrol at 7, 14, and 21 days after treatment was 78.4, 69.5, 60.5%, respectively, in crude extract treatment plots and 77.1, 70.2, and 57.3% in fresh cells culture of SB177 treatment plots (Table 8). In comparison, the biocontrol efficacy of Jinggangmycin and Kresoxim-methyl treatment plots at 7, 14, and 21 days after treatment was 67.1, 57.9, and 48.1%; and 63.8, 51.9, and 48.5% (Table 8), respectively.
A similar trend was observed in the 2014 filed experiment and both the crude extract from NF0919 culture filtrate and fresh cells culture of SB177 showed better disease control efficacy than Jinggangmycin or Kresoxim-methyl (P < 0.1) (Table 9). The collective results from the 2-year field experiment suggested that the crude extract of NF0919 and fresh cells of SB177 provided better disease control efficacy than the fungicides Jinggangmycin or Kresoxim-methyl.
Discussion
Rice sheath blight is a serious disease in China [54]. Application biocontrol agent could greatly reduce the use of fungicides and thereby reduce pollution of the environment. Based on these above point of views, and looking forward to screening potential biocontrol microbial, we carried out this research.
Previous studies have indicated that cowdung (CD) microflora showing diversity, and normally contain abundant number of bacilli, lactobacilli, cocci, some identified and unidentified fungi and yeasts [37]. Ware et al. [58] reported that the lower part of the gut of the cow contains various microorganisms including Lactobacillus plantarum, L. casei, L. acidophilus, B. subtilis, Enterococcus diacetylactis. Other than these, the rumen of the cow contains various species of Bacillus and Bifidobacterium and yeasts (commonly Saccharomyces cerevisiae) for better rumen fermentation [27]. Under normal conditions, aged CD gets invaded with several soil contaminants such as bacteria, fungi, Trichoderma and actinomycetes [37]. Swain and Ray [52] reported that some bacteria (isolated from CD), B. subtilis CM1-CM5 had the ability to inhibit the in vitro growth of fungi.
In the current study, we isolated a new Bacillus strain SB177 from CD microflora and identified it as a strain of B. amyloliquefaciens. In dual-culture assays, mycelial growth of R. solani was strongly inhibited by SB177. Further study revealed that the inhibited mycelia of R. solani cannot infect the rice leaf again (paper in preparation). Our data indicate that the control efficacy of treatment with SB177 was 77.1% in 2013 and 94.2% in 2014 after treatment 7 days (Tables 8, 9) in field test. Furthermore, studies in our experiment have shown that all these antagonistic Bacillus spp (B. cereus JR08, B. amyloliquefaciens Mg116, B. amyloliquefaciens RM09, and B. amyloliquefaciens SB177) were thermotolerant (up to 60 °C), which may be useful in producing commercial products.
Although the group of fluorescent pseudomonas included the species P. fluorescens PF1, PB2, PfALR1, pfMDU2, and P. aureofaciens have been reported to be the most frequently beneficial and antifungal bacteria [38–40, 47, 56], due to the characteristics of the fluorescent pseudomonas (short shelf-life of formulation), there were almost no commercial products to be used in control RSB so far.
For biocontrol applications, Bacillus spp. was commonly used, because they can produce endospores, and can be well compatible with commercial products in factories. Almost all of the biocontrol products of RSB were Bacillus spp. included the species B. cereus, B. subtilis Bs-916, B. megaterium, B. subtilis NJ-18 and B. subtilis MBI 600 [9, 10, 26, 30, 68]. Moreover, B. subtilis formulations were used to control RSB in combination with a fungicide and antibiotic (Jianggangmycin), or as single bacteria formulations [59, 60, 63].
Understanding the mechanism of control could help us improve the level and consistency of control. The modes of action reported for bacterial antagonists of fungal pathogens are antibiosis, nutrient depletion around the sites of pathogen penetration, hyper-parasitism with release of cell wall degrading enzymes, stimulation of the plant’s defense capacity and induction of systemic resistance (ISR) [1, 3, 18, 29, 33, 36, 41, 45, 55].
In our study, the antagonism of selected bacterial isolates was found to involve the production of antifungal compounds. In dual-culture experiments, the wide inhibition halos produced by the bacterial isolates SB177 toward the fungal mycelia of R. solani suggest the release of bacterial-diffusing antimicrobial compounds. Although our in vitro data indicate that B. subtilis strain SB177 inhibits R. solani by means of antibiosis, we have no direct evidence that antibiosis is the mode of action in the field. Given that strain SB177 provided high levels of control in the field, however, the mode of action in the field warrants investigation. Further investigations will also be aimed at identifying and characterizing such bioactive molecules.
Bioactive compounds, because of their natural origin, are biodegradable and do not leave toxic residues or byproducts to contaminate the environment, whereas commercial fungicides pose severe toxicity to humans, plants and animals [34]. There were lots of reports about the use of Streptomyces spp. to control RSB [2, 6, 21, 31, 43, 44, 50, 62, 65]. From the beginning of 1970s, China and Japan were all used the metabolic products validamycin (purificated from the metabolites of S. hygroscopicus var. limoneus nov. var. in Japan and S. hygroscopicus var. jinggangensis nov. var in China) to control RSB [8, 21]. Some reports have clarified that the efficacy of validamycin (also named jinggangmycin in China) to RSB was decreased [23, 61, 66]. Therefore, screening and discovery of potential candidate isolates of actinomycetes appears to be of great significance.
In our present study, cowdung-associated Streptomyces species found was not previously reported as R. solani antagonists. It was a new isolate of Streptomyces spp., labeled strain NF0919, was selected on the basis of its antifungal activity to RSB in vivo and in vitro conditions. The strain was identified as a strain of S. cochorusii. A 2-year field experiment revealed that 167.0 mg a.i. l−1 of the crude extract from NF0919 culture filtrate effectively reduced the development of RSB from 78.4 to 98.1%. The biocontrol efficacy was higher than the 63.1–79.7% efficacy of 300.0 mg a.i. l−1 of the fungicide Kresoxim-methyl and the 67.1–83.8% efficacy of 500.0 mg a.i. l−1 of the antibiotic Jinggangmycin in controlling the disease 7 days after treatment. It was important to note that the recommended dosage of Jinggangmycin in China was only 160.0 mg a.i. l−1 for field control. The mechanism of Jinggangmycin to R. solani. is still unclear. One kind of mechanism of Jinggangmycin was the inhibition of the trehalose enzyme of R. solani [24]. Jinggangmycin have no direct inhibitory effect on R. solani in vitro test [32]. However, the metabolites of our study have direct inhibition to the mycelium of R. solani. As a result, it seems to conclude that the metabolic product in our study was different from the Jinggangmycin and may be a novel type of antibiotic.
Our results suggested that strains NF0919 and SB177 may be developed as a biocontrol resource for RSB control. To our knowledge, this is the first report of the use of B. amyloliquefaciens and S. cochorusii as biocontrol resources for RSB. Further research will be carried out to establish stable formulations, interpret the mechanism of the biocontrol agents, and identify the molecular structural formula of secondary metabolites.
References
Ahimoua, F., Jacquesb, P., & Deleua, M. (2000). Surfactin and iturin A effects on Bacillus subtilis surface hydrophobicity. Enzyme and Microbial Technology, 27, 749–754.
Asano, N., Yamaguchi, T., & Kameda, Y. (1987). Effect of Vlidamycins on glycohydrolases of Rhizoctonia solani. Journal of Antibiotics, 40, 526–532.
Bakker, P. A. H. M., Pieterse, C. M. J., & Van Loon, L. C. (2007). Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology, 97, 239–243.
Berg, G., Krechel, A., Ditz, M., Sikora, R. A., Ulrich, A., & Hallmann, J. (2005). Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiology Ecology, 51, 215–229.
Bonmann, J. M., Khush, G. S., & Nelson, R. J. (1992). Breeding rice for resistance to pests. Annual review of Phytopathology, 30, 507–528.
Boukaew, S., & Prasertsan, P. (2014). Suppression of rice sheath blight disease using a heat stable culture filtrate from Streptomyces philanthi RM-1-138. Crop Protection, 61, 1–10.
Bozzola, J. J., & Russell, L. D. (1999). Electron microscopy principles and techniques for biologists (2nd ed.). Sudbury, MA: Jones and Bartlett publishers.
Chen, X. L., Fang, X., & Shen, Y. C. (2010). Mechanism, resistance and security of Jinggangmycin against Rhizoctonia solani. Agrochemicals, 7, 481–483. (mini review in Chinese, with English abstract).
Chen, Z. Y., Liu, Y. F., & Lu, F. (2004). Study on key technology in the industrialized production of Bacillus subtilis Bs-916, the rice sheath blight control agent. Acta Phytophylacica Sinica, 31, 230–234. (in Chinese, with English abstract).
Chen, Z. Y., Xue, Z. G., Lu, F., Liu, Y. F., & Chen, Y. L. (2000). On antagonism against Rhizoctonia solani of culture solution of strain B-916 and constituent of its angifungal substance. Jiangsu Journal of Agricultural Sciences, 16, 148–152. (in Chinese, with English abstract).
Chernin, L., Ismailov, Z., Haran, S., & Chet, I. (1995). Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Applied and Environment Microbiology, 61, 1720–1726.
Chin, K. M., & Bhandhufalck, A. (1990). The importance of crop growth stages for determining the application timing of disease control agents on rice. In B. T. Grayson, M. B. Green, & L. G. Copping (Eds.), Pest management in rice (pp. 131–142). London: Elsevier.
Damicone, J. P., Patel, M. V., & Moore, W. F. (1993). Density of sclerotia of Rhizoctonia solani and incidence of sheath blight in rice fields in Mississippi. Plant Disease, 77, 257–260.
Das, K., Doley, R., & Mukherjee, A. K. (2004). Purification and characterization of a thermostable, alkaliphilic, extracellular alpha-amylase from Bacillus subtilis DM-03, a strain isolated from the traditional fermented food of India. Biotechnology and Applied Biochemistry, 40, 291–298.
Garrity, G. (2001). Bergey’s manual of systematic bacteriology (2nd ed.). New York, NY: Springer.
Ghose, T. K. (1987). Measurement of cellulase activities. Pure and Applied Chemistry, 59, 257–268.
Gottlieb, D. (1961). An evolution of criteria and procedures used in the description and characterization of Streptomyces, a cooperative study. Applied Microbiology, 9, 55–60.
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., & Lorito, M. (2004). Trichoderma species—opportunistic, avirulent plant symbionts. Nature Reviews Microbiology, 2, 43–56.
Hu, X. R., Xu, W. Y., & Lu, W. C. (2006). The sensitivity of rice sheath blight fungi (Rhizoctonia solani) to Jinggangmycin in Fujian province of China. Chinese Agricultural Science Bulletin, 22, 160–162. (in Chinese, with English abstract).
IRRI. (1980). Standard evaluation system for rice. Philippines: International Rice Research Institute.
Iwasa, T., Yamamoto, H., & Shibata, M. (1970). Studies on validamycins, new antibiotics. I: Streptomyces hygroscopicus var. limoneus nov. var., validamycin-producing organism. Japanese Journal of Antibiotics, 23, 595–602.
Jiang, B. G., & Zhao, B. (2005). Screening and identification of the bacteria which have high efficiency on resolving phosphorus and potassium and in nitrogen fixation. Journal of Huazhong Agricultural University, 24, 43–48. (in Chinese, with English abstract).
Jin, M. S., & Jiang, W. L. (1997). Detection of resistance to Jinggangmycin in rice sheath blight, Thanatephorus cucumeris (Frank) Donk. From Zhejiang province. Acta Agriculturae Zhejiangensis, 9, 127–130. (in Chinese, with English abstract).
Joaìo, A. J. (1997). Trehalases and trehalose hydrolysis in fungi. FEMS Microbiology Letters, 154, 165–171.
Kucey, R. M. N. (1983). Phosphate-solubilizing bacteria and fungi in various cultivated and virgin Alberta soils. Canadian Journal of Soil Science, 63, 671–678.
Kumar, K. V., Yellareddygari, S. K. R., Reddy, M. S., Kloepper, J. W., Lawrence, K. S., Zhou, X. G., et al. (2012). Efficacy of Bacillus subtilis MBI 600 against sheath blight caused by Rhizoctonia solani and on growth and yield of rice. Rice Science, 19, 55–63.
Kung, J. L. (2004). Accessed on 10 February 2005. A direct fed microbes and enzyme for dairy cows. www.das.psu.edu/dairynutrition/documents/kung.pdf.
Lee, F. N., & Rush, M. C. (1983). Rice sheath blight: A major rice disease. Plant Disease, 67, 829–832.
Leifert, C., Li, H., Chidburee, S., Hampson, S., Workman, S., Sigee, D., et al. (1995). Antibiotic production and biocontrol activity by Bacillus subtilis CL27 and Bacillus pumilus CL45. Journal of Applied Bacteriology, 78, 97–108.
Li, H. R., Xiao, J. G., & Yan, S. Q. (1993). Biological control of rice sheath blight by Bacillus cereus. Acta Phytopathologica Sinica, 23, 101–105. (in Chinese, with English abstract).
Liao, Y. Q., Wei, Z. H., Bai, L. Q., Deng, Z. X., & Zhong, J. J. (2009). Effect of fermentation temperature on validamycin A production by Streptomyces hygroscopicus 5008. Journal of Biotechnology, 142, 271–274.
Liu, H. M., & Lu, G. Z. (2008). Action mechanisms of validamycin a controlling rice sheath blight. Dissertation for Master Degree of Fujiang agricultural University, Fujiang, China.
Liu, Y. F., Chen, Z. Y., Ng, T. B., Zhang, J., Zhou, M. G., Song, F. P., et al. (2007). Bacisubin, an antifungal protein with ribonuclease and hemagglutinating activities from Bacillus subtilis strain B-916. Peptides, 28, 553–559.
Mantovani, A., Maranghi, F., Ricciardi, C., Marci, C., Stazi, A. V., Attias, L., et al. (1998). Developmental toxicity of carbendazim: Comparison of no observed-adverse effect level and bench mark dose approach. Food and Chemical Toxicology, 36, 37–45.
Meng, Q. Z., Liu, Z. H., Wang, H. Y., Zhang, S. S., & Wei, S. H. (2001). Research progress in rice sheath blight. Journal of Shenyang Agricultural University, 32, 376–381. (in Chinese, with English abstract).
Montealegre, J. R., Reyes, R., Perez, L. M., Herrera, R., Silva, P., & Besoain, X. (2003). Selection of bio-antagonistic bacteria to be used in biological control of Rhizoctonia solani in tomato. Electronic Journal of Biotechnology, 6, 115–127.
Muhammad, S., & Amusa, N. A. (2003). In vitro inhibition of growth of some seedling blight inducing pathogens by compost inhibiting microbes. African Journal of Biotechnology, 2, 161–164.
Nagarajkumar, M., Bhaskaran, R., & Velazhahan, R. (2004). Involvement of secondary metabolites and extracellular lytic enzymes produced by Pseudomonas fluorescens in inhibition of Rhizoctonia solani, the rice sheath blight pathogen. Microbiological Research, 159, 73–81.
Nagarajkumar, M., Jayaraj, J., Muthukrishnan, S., Bhaskaran, R., & Velazhahan, R. (2005). Detoxification of oxalic acid by Pseudomonas fluorescens strain pfMDU2: Implications for the biological control of rice sheath blight caused by Rhizoctonia solani. Microbiological Research, 160, 291–298.
Nandakumar, R., Babu, S., Viswanathan, R., Sheela, J., Raguchander, T., & Samiyappan, R. (2001). A new bio-formulation containing plant growth promoting rhizobacterial mixture for the management of sheath blight and enhanced grain yield in rice. BioControl, 46, 493–510.
Pal Bais, H., Fall, R., & Vivanco, J. M. (2004). Biocontrol of Bacillus subtilis against infection of arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiology, 134, 307–319.
Pengnoo, A., Kusonwiriyawong, C., Nilratana, L., & Kanjanamaneesathian, M. (2000). Greenhouse and field trials of the bacterial antagonists in pellet formulations to suppress sheath blight of rice caused by Rhizoctonia solani. BioControl, 45, 245–256.
Prabavathy, V. R. (2005). Isolation, purification and characterization of antimicrobial metabolites produced by Streptomyces sp. and evaluation against blast and sheath blight diseases of rice. Ph.D. Thesis, University of Madras, Chennai, India.
Prabavathy, V. R., Mathivanan, N., & Murugesan, K. (2006). Control of blast and sheath blight diseases of rice using antifungal metabolites produced by Streptomyces sp. PM5. Biological Control, 39, 313–319.
Raaijmakers, J. M., Vlami, M., & de Souza, J. T. (2002). Antibiotic production by bacterial biocontrol agents. Antonie van Leeuwenhoek, 81, 537–547.
Rabindran, R., & Vidhyasekaran, P. (1996). Development of a formulation of Pseudomonas fluorescens PfALR2 for management of rice sheath blight. Crop Protection, 15, 715–721.
Radja, C. R., Nandakumar, R., Kandan, A., Suresh, S., Bharathi, M., Raguchander, T., et al. (2002). Pseudomonas fluorescens based bio-formulation for the management of sheath blight disease and leaffolder insect in rice. Crop Protection, 21, 671–677.
Reissing, W. H., Heinrichs, E. A., Litsinger, J. A., Moody, K., Fieldeer, L., Mew, T. W., et al. (1986). Illustrated guide to integrated pest management in rice in tropical Asia. Los Baños, Laguna: International Rice Research Institute.
Shanmugam, V., Siram, S., & Babu, S. (2001). Purification and characterization of an extracellular alpha-glucosidase protein from Trichoderma viride thick degrades a phytotoxin associated with sheath blight disease in rice. Journal of Applied Microbiology, 90, 320–329.
Shen, Y. C. (1988). Research and development of agro-antibiotic, validamycin. Chinese Journal of Antibiotics, 6, 58–61. (in Chinese, with English abstract).
Shin, S. H., Lim, Y., Lee, S. E., Yang, N. W., & Rhee, J. H. (2001). CAS agar diffusion assay for the measurement of siderophores in biological fluids. Journal of Microbiological Methods, 44, 89–95.
Swain, M. R., & Ray, R. C. (2009). Biocontrol and other beneficial activities of Bacillus subtilis isolated from cowdung microflora. Microbiological Research, 164, 121–130.
Tang, J. B., Ma, B. T., Wang, L. X., Li, P., Zheng, A. P., & Chen, H. (2002). Biological control of rice sheath blight with Trichoderma and Trichoderma-like. Chinese Journal of Rice Science, 16, 63–66. (in Chinese, with English abstract).
Teng, P. S., Torres, C. Q., Nuque, F. L., & Calvero, S. B. (1990). Current knowledge on crop losses in tropical rice: Crop loss assessment in rice (pp. 39–53). Manila: International Rice Research Institute.
Van der Ent, S., Verhagen, B. W. M., Van Doorn, R., Bakker, D., Verlaan, M. G., Pel, M. J. C., et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiology, 146, 1293–1304.
Vidhyasekaran, P., & Muthamilan, M. (1999). Evaluation of powder formulations of Pseudomonas fluorescens Pf1 for control of rice sheath blight. Biocontrol Science and Technology, 9, 67–74.
Wang, Y. L., Shen, Y., & Xu, T. (2000). Study on Trichoderma harzianum strains to control rice sheath blight. Acta Phytophylacica Sinica, 27, 97–101. (in Chinese, with English abstract).
Ware, D. R., Read, P. L., & Manfredi, E. T. (1998). Lactation performance of two large diary herds fed Lactobacillus acidophilus strain BT1386. Journal of Dairy Science, 71(Suppl 1), 219–222.
Wiwattanapatapee, R., Chumthong, A., Pengnoo, A., & Kanjanamaneesathian, M. (2007). Effervescent fast-disintegrating bacterial formulation for biological control of rice sheath blight. Journal of Controlled Release, 119, 229–235.
Wiwattanapatapee, R., Pengnoo, A., Kanjanamaneesathian, M., Matchavanich, W., Nilratana, L., & Jantharangsri, A. (2004). Floating pellets containing bacterial antagonist for control sheath blight of rice: Formulations, viability and bacterial release studies. Journal of Controlled Release, 95, 455–462.
Wu, J., Xi, Y. D., Li, H. H., Wang, X. L., & Peng, H. X. (2015). Monitoring of resistance of Thenatephorus cucumeris to Jinggangmysin in Sichuan. Southwest China Journal of Agricultural Sciences, 28, 2501–2504. (in Chinese, with English abstract).
Xu, L. H., Li, Q. R., & Jiang, C. L. (1996). Diversity of soil actinomycetes in Yunnan, China. Applied Environmental Microbiology, 62, 244–248.
Yang, D. J., Wang, B., Wang, J. X., Chen, Y., & Zhou, M. G. (2009). Activity and efficacy of Bacillus subtilis strain NJ-18 against rice sheath blight and Sclerotinia stem rot of rape. Biological Control, 51, 61–65.
Yang, J. H., Liu, H. X., Zhu, G. M., Pan, Y. L., Xu, L. P., & Guo, J. H. (2008). Diversity analysis of antagonists from rice-associated bacteria and their application in biocontrol of rice diseases. Journal of Applied Microbiology, 104, 91–104.
Yu, Y., Bai, L., & Minagawa, K. (2005). Gene cluster responsible for Validamycin biosynthesis in Streptomyces hygroscopicus subsp. Jinggangensis 5008. Applied and Environment Microbiology, 71, 5066–5076.
Zhang, S., Xu, W. X., Xue, Y. G., Chen, X. X., & Wang, J. R. (1995). A preliminary study on the sensitivity of rice sheath blight fungi (Rhizoctonia solani) to Jinggangmycin in Zhengzhou suburbs. Chinese Journal of Biological Control, 11, 171–173. (in Chinese, with English abstract).
Zhen, A. P., Li, P., Wang, L. X., Ma, B. T., & Wang, X. Q. (2001). Screening of the antagonistic bacteria against rice sheath blight and study on the antibiosis. Southwest China Journal of Agricultural Science, 14, 78–81. (in Chinese, with English abstract).
Zhen, A. P., Li, P., Wang, S. Q., & Shun, H. Q. (2003). Screening and taxonomy of antagonistic strain B34 against Thanatephorus cucumeris. Acta Phytopathologica Sinica, 33, 81–85. (in Chinese, with English abstract).
Zou, J. H., Pan, X. B., Chen, Z. X., Xu, J. Y., Lu, J. F., Zhai, W. X., et al. (2000). Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.). Theoretical and Applied Genetics, 101, 569–573.
Acknowledgements
This research was supported by a Grant-in-Aid for science research from the Independent innovation Foundation (cx(12)5067), and the Science and Technological Project (NY2015019, BE2015364) of Jiangsu province. We thank the anonymous reviewers for critical comments and suggestions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jing-Hui Yang and Wen-Wen Zhang contributed equally to this study and are regarded as joint first authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Yang, JH., Zhang, WW., Zhuang, YQ. et al. Biocontrol activities of bacteria from cowdung against the rice sheath blight pathogen. J Plant Dis Prot 124, 131–141 (2017). https://doi.org/10.1007/s41348-017-0080-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-017-0080-1