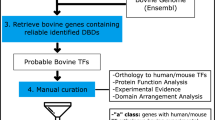
Abstract
The basic helix–loop–helix (bHLH) transcription factors (TFs) form a huge superfamily and play crucial roles in many essential developmental, genetic, and physiological–biochemical processes of eukaryotes. In total, 109 putative bHLH TFs were identified and categorized successfully in the genomic databases of cattle, Bos Taurus, after removing redundant sequences and merging genetic isoforms. Through phylogenetic analyses, 105 proteins among these bHLH TFs were classified into 44 families with 46, 25, 14, 3, 13, and 4 members in the high-order groups A, B, C, D, E, and F, respectively. The remaining 4 bHLH proteins were sorted out as ‘orphans.’ Next, these 109 putative bHLH proteins identified were further characterized as significantly enriched in 524 significant Gene Ontology (GO) annotations (corrected P value ≤ 0.05) and 21 significantly enriched pathways (corrected P value ≤ 0.05) that had been mapped by the web server KOBAS 2.0. Furthermore, 95 bHLH proteins were further screened and analyzed together with two uncharacterized proteins in the STRING online database to reconstruct the protein–protein interaction network of cattle bHLH TFs. Ultimately, 89 bHLH proteins were fully mapped in a network with 67 biological process, 13 molecular functions, 5 KEGG pathways, 12 PFAM protein domains, and 25 INTERPRO classified protein domains and features. These results provide much useful information and a good reference for further functional investigations and updated researches on cattle bHLH TFs.



Similar content being viewed by others
References
Arnold DR, Bordignon V, Lefebvre R, Murphy BD, Smith LC (2006a) Somatic cell nuclear transfer alters peri-implantation trophoblast differentiation in bovine embryos. Reproduction 132:279–290
Arnold DR, Lefebvre R, Smith LC (2006b) Characterization of the placenta specific bovine mammalian achaete scute-like homologue 2 (Mash2) gene. Placenta 27:1124–1131
Atchley WR, Fitch WM (1997) A natural classification of the basic helix–loop–helix class of transcription factors. Proc Natl Acad Sci USA 94:5172–5176
Atchley WR, Terhalle W, Dress A (1999) Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol E 48:501–516
Baudier J, Bergeret E, Bertacchi N, Weintraub H, Gagnon J, Garin J (1995) Interactions of myogenic bHLH transcription factors with calcium-binding calmodulin and S100a (alpha alpha) proteins. Biochem 34:7834–7846
Ben-Shoshan J, Maysel-Auslender S, Luboshits G, Barshack I, Polak-Charcon S, Tzahor E, Keren G, George J (2009) Hypoxia-inducible factor-1alpha and—2alpha additively promote endothelial vasculogenic properties. J Vasc Res 46:299–310
Berkes CA, Tapscott SJ (2005) MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16:585–595
Breen GA, Jordan EM (1997) Regulation of the nuclear gene that encodes the alpha-subunit of the mitochondrial F0F1-ATP synthase complex. Activation by upstream stimulatory factor 2. J Biol Chem 272:10538–10542
Brennan KM, Vella KR, Good DJ (2006) Genetic analysis of NHLH2 and its putative role in bovine body weight control. Anim Genet 37 Suppl 1:24–27
Calomme C, Nguyen TL, de Launoit Y, Kiermer V, Droogmans L, Burny A, Van Lint C (2002) Upstream stimulatory factors binding to an E box motif in the R region of the bovine leukemia virus long terminal repeat stimulates viral gene expression. J Biol Chem 277:8775–8789
Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153:1398–1412
Chengxiao Z, Ze Y (2015) Biological function and molecular mechanism of Twist2. Yi Chuan 37:17–24
Comai G, Tajbakhsh S (2014) Molecular and cellular regulation of skeletal myogenesis. Curr Top Dev Biol 110, 1–73
Dang CW, Wang Y, Chen KP, Yao Q, Zhang DB, Guo M (2011) The basic helix-loop-helix transcription factor family in the pea aphid, Acyrthosiphon pisum. J Insect Sci 11:84
Datta TK, Rajput SK, Wee G, Lee K, Folger JK, Smith GW (2015) Requirement of the transcription factor USF1 in bovine oocyte and early embryonic development. Reproduction 149:203–212
Dechesne CA, Wei Q, Eldridge J, Gannoun-Zaki L, Millasseau P, Bougueleret L, Caterina D, Paterson BM (1994) E-box- and MEF-2-independent muscle-specific expression, positive autoregulation, and cross-activation of the chicken MyoD (CMD1) promoter reveal an indirect regulatory pathway. Mol Cell Biol 14:5474–5486
Deng Q, Gao Y, Jiang H, Chen CZ, Li CH, Yu WL, Chen X, Zhang JB (2015) Association of a hypoxia-inducible factor-3alpha gene polymorphism with superovulation traits in Changbaishan black cattle. Genet Mol Res 14:14539–14547
Dolt KS, Karar J, Mishra MK, Salim J, Kumar R, Grover SK, Qadar PM (2007a) Transcriptional downregulation of sterol metabolism genes in murine liver exposed to acute hypobaric hypoxia. Biochem Biophys Res Commun 354:148–153
Dolt KS, Mishra MK, Karar J, Baig MA, Ahmed Z, Pasha MA (2007b) cDNA cloning, gene organization and variant specific expression of HIF-1 alpha in high altitude yak (Bos grunniens). Gene 386:73–80
Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, Weinstock GM, Adelson DL, Eichler EE, Elnitski L, Guigo R, Hamernik DL, Kappes SM, Lewin HA, Lynn DJ, Nicholas FW, Reymond A, Rijnkels M, Skow LC, Zdobnov EM, Schook L, Womack J, Alioto T, Antonarakis SE, Astashyn A, Chapple CE, Chen HC, Chrast J, Camara F, Ermolaeva O, Henrichsen CN, Hlavina W, Kapustin Y, Kiryutin B, Kitts P, Kokocinski F, Landrum M, Maglott D, Pruitt K, Sapojnikov V, Searle SM, Solovyev V, Souvorov A, Ucla C, Wyss C, Anzola JM, Gerlach D, Elhaik E, Graur D, Reese JT, Edgar RC, McEwan JC, Payne GM, Raison JM, Junier T, Kriventseva EV, Eyras E, Plass M, Donthu R, Larkin DM, Reecy J, Yang MQ, Chen L, Cheng Z, Chitko-McKown CG, Liu GE, Matukumalli LK, Song J, Zhu B, Bradley DG, Brinkman FS, Lau LP, Whiteside MD, Walker A, Wheeler TT, Casey T, German JB, Lemay DG, Maqbool NJ, Molenaar AJ, Seo S, Stothard P, Baldwin CL, Baxter R, Brinkmeyer-Langford CL, Brown WC, Childers CP, Connelley T, Ellis SA, Fritz K, Glass EJ, Herzig CT, Iivanainen A, Lahmers KK, Bennett AK, Dickens CM, Gilbert JG, Hagen DE, Salih H, Aerts J, Caetano AR, Dalrymple B, Garcia JF, Gill CA, Hiendleder SG, Memili E, Spurlock D, Williams JL, Alexander L, Brownstein MJ, Guan L, Holt RA, Jones SJ, Marra MA, Moore R, Moore SS, Roberts A, Taniguchi M, Waterman RC, Chacko J, Chandrabose MM, Cree A, Dao MD, Dinh HH, Gabisi RA, Hines S, Hume J, Jhangiani SN, Joshi V, Kovar CL, Lewis LR, Liu YS, Lopez J, Morgan MB, Nguyen NB, Okwuonu GO, Ruiz SJ, Santibanez J, Wright RA, Buhay C, Ding Y, Dugan-Rocha S, Herdandez J, Holder M, Sabo A, Egan A, Goodell J, Wilczek-Boney K, Fowler GR, Hitchens ME, Lozado RJ, Moen C, Steffen D, Warren JT, Zhang J, Chiu R, Schein JE, Durbin KJ, Havlak P, Jiang H, Liu Y, Qin X, Ren Y, Shen Y, Song H, Bell SN, Davis C, Johnson AJ, Lee S, Nazareth LV, Patel BM, Pu LL, Vattathil S, Williams RJ, Curry S, Hamilton C, Sodergren E, Wheeler DA, Barris W, Bennett GL, Eggen A, Green RD, Harhay GP, Hobbs M, Jann O, Keele JW, Kent MP, Lien S, McKay SD, McWilliam S, Ratnakumar A, Schnabel RD, Smith T, Snelling WM, Sonstegard TS, Stone RT, Sugimoto Y, Takasuga A, Taylor JF, Van Tassell CP, Macneil MD, Abatepaulo AR, Abbey CA, Ahola V, Almeida IG, Amadio AF, Anatriello E, Bahadue SM, Biase FH, Boldt CR, Carroll JA, Carvalho WA, Cervelatti EP, Chacko E, Chapin JE, Cheng Y, Choi J, Colley AJ, de Campos TA, De Donato M, Santos IK, de Oliveira CJ, Deobald H, Devinoy E, Donohue KE, Dovc P, Eberlein A, Fitzsimmons CJ, Franzin AM, Garcia GR, Genini S, Gladney CJ, Grant JR, Greaser ML, Green JA, Hadsell DL, Hakimov HA, Halgren R, Harrow JL, Hart EA, Hastings N, Hernandez M, Hu ZL, Ingham A, Iso-Touru T, Jamis C, Jensen K, Kapetis D, Kerr T, Khalil SS, Khatib H, Kolbehdari D, Kumar CG, Kumar D, Leach R, Lee JC, Li C, Logan KM, Malinverni R, Marques E, Martin WF, Martins NF, Maruyama SR, Mazza R, McLean KL, Medrano JF, Moreno BT, More DD, Muntean CT, Nandakumar HP, Nogueira MF, Olsaker I, Pant SD, Panzitta F, Pastor RC, Poli MA, Poslusny N, Rachagani S, Ranganathan S, Razpet A, Riggs PK, Rincon G, Rodriguez-Osorio N, Rodriguez-Zas SL, Romero NE, Rosenwald A, Sando L, Schmutz SM, Shen L, Sherman L, Southey BR, Lutzow YS, Sweedler JV, Tammen I, Telugu BP, Urbanski JM, Utsunomiya YT, Verschoor CP, Waardenberg AJ, Wang Z, Ward R, Weikard R, Welsh TJ, White SN, Wilming LG, Wunderlich KR, Yang J, Zhao FQ (2009) The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324:522–528
Ferre-D’Amare AR, Prendergast GC, Ziff EB, Burley SK (1993) Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363:38–45
Fraisl P, Mazzone M, Schmidt T, Carmeliet P (2009) Regulation of angiogenesis by oxygen and metabolism. Dev Cell 16:167–179
Gohil K, Brooks GA (2012) Exercise tames the wild side of the Myc network: a hypothesis. Am J Physiol Endocrinol Metab 303:E18–E30
Hara S, Kobayashi C, Imura N (1999) Molecular cloning of cDNAs encoding hypoxia-inducible factor (HIF)-1alpha and—2alpha of bovine arterial endothelial cells. Biochim Biophys Acta 1445:237–243
Harvey AJ, Kind KL, Thompson JG (2004a) Effect of the oxidative phosphorylation uncoupler 2,4-dinitrophenol on hypoxia-inducible factor-regulated gene expression in bovine blastocysts. Reprod Fertil Dev 16:665–673
Harvey AJ, Kind KL, Pantaleon M, Armstrong DT, Thompson JG (2004b) Oxygen-regulated gene expression in bovine blastocysts. Biol Reprod 71:1108–1119
Harvey AJ, Kind KL, Thompson JG (2007) Regulation of gene expression in bovine blastocysts in response to oxygen and the iron chelator desferrioxamine. Biol Reprod 77:93–101
Heisler MG, Atkinson A, Bylstra YH, Walsh R, Smyth DR (2001) SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128:1089–1098
Hu J, Peng XZ (2007) [Research advances in nervous system-specific transcription factor, Nhlh2]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 29:436–440
Hudson NJ, Lyons RE, Reverter A, Greenwood PL, Dalrymple BP (2013) Inferring the in vivo cellular program of developing bovine skeletal muscle from expression data. Gene Expr Patterns 13:109–125
Hyun Y, Lee I (2006) KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol 61:283–296
Jung H, Choe Y, Kim H, Park N, Son GH, Khang I, Kim K (2003) Involvement of CLOCK:BMAL1 heterodimer in serum-responsive mPer1 induction. Neuroreport 14:15–19
Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, Szczyglowski K (2009) Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol 151:1175–1185
Khanbabaei H, Teimoori A, Mohammadi M (2016) The interplay between microRNAs and Twist1 transcription factor: a systematic review. Tumour Biol 37:7007–7019
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol E 33:1870–1874
Ledent V, Vervoort M (2001) The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res 11:754–770
Ledent V, Paquet O, Vervoort M (2002) Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol 3:H30
Lee JE (1997) Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol 7:13–20
Lee S, Lee S, Yang KY, Kim YM, Park SY, Kim SY, Soh MS (2006) Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol 47:591–600
Leow CC, Polakis P, Gao WQ (2005) A role for Hath1, a bHLH transcription factor, in colon adenocarcinoma. Ann NY Acad Sci 1059:174–183
Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141:1167–1184
Li HJ, Ray SK, Singh NK, Johnston B, Leiter AB (2011) Basic helix-loop-helix transcription factors and enteroendocrine cell differentiation. Diabetes Obes Metab 13 Suppl 1:5–12
Liu W (2015) Genome-wide identification, classification and functional analyses of the bHLH transcription factor family in the pig, Sus scrofa. Mol Genet Genomics 290:1415–1433
Liu W, Chen D (2013) Phylogeny, functional annotation, and protein interaction network analyses of the Xenopus tropicalis basic helix-loop-helix transcription factors. Biomed Res Int 2013, 145037
Liu W, Li F (2015) Identification and Bioinformatics Analyses of the Basic Helix-loop-helix Transcription Factors in Xenopus laevis. Pak J Biol Sci 18:149–165
Liu WY, Zhao CJ (2010) Genome-wide identification and analysis of the chicken basic helix-loop-helix factors. Comp Funct Genomics, 682095
Liu W, Zhao C (2011) Molecular phylogenetic analysis of zebra finch basic helix-loop-helix transcription factors. Biochem Genet 49:226–241
Liu A, Wang Y, Dang C, Zhang D, Song H, Yao Q, Chen K (2012) A genome-wide identification and analysis of the basic helix-loop-helix transcription factors in the ponerine ant, Harpegnathos saltator. BMC Evol Biol 12:165
Liu A, Wang Y, Zhang D, Wang X, Song H, Dang C, Yao Q, Chen K (2013) Classification and evolutionary analysis of the basic helix-loop-helix gene family in the green anole lizard, Anolis carolinensis. Mol Genet Genomics 288:365–380
Liu XT, Wang Y, Wang XH, Tao XF, Yao Q, Chen KP (2014) A genome-wide identification and classification of basic helix-loop-helix genes in the jewel wasp, Nasonia vitripennis (Hymenoptera: Pteromalidae). Genome 57:525–536
Liu Q, Wu Y, Yoshizawa T, Yan X, Morohashi S, Seino H, Kato Y, Kijima H (2016) Basic helix-loop-helix transcription factor DEC2 functions as an anti-apoptotic factor during paclitaxel-induced apoptosis in human prostate cancer cells. Int J Mol Med 38:1727–1733
Masoud GN, Li W (2015) HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B 5:378–389
Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20:429–440
Miraoui H, Marie PJ (2010) Pivotal role of Twist in skeletal biology and pathology. Gene 468:1–7
Moncaut N, Rigby PW, Carvajal JJ (2013) Dial M(RF) for myogenesis. Febs J 280:3980–3990
Mulvaney J, Dabdoub A (2012) Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J Assoc Res Otolaryngol 13:281–293
Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, Et A (1989) Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58:537–544
Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH (1994) Structure and function of helix-loop-helix proteins. Biochim Biophys Acta 1218:129–135
Nicholas K B NHBD (1996) Genedoc: Analysis and Visualization of Genetic Variation. Embnew News 4:28–30
Nilsson I, Shibuya M, Wennstrom S (2004) Differential activation of vascular genes by hypoxia in primary endothelial cells. Exp Cell Res 299:476–485
Noguchi H, Ueda M, Matsumoto S, Kobayashi N, Hayashi S (2007) BETA2/NeuroD protein transduction requires cell surface heparan sulfate proteoglycans. Hum Gene Ther 18:10–17
Park YS, Kim G, Jin YM, Lee JY, Shin JW, Jo I (2016) Expression of angiopoietin-1 in hypoxic pericytes: regulation by hypoxia-inducible factor-2alpha and participation in endothelial cell migration and tube formation. Biochem Biophys Res Commun 469:263–269
Patel D, Morton DJ, Carey J, Havrda MC, Chaudhary J (2015) Inhibitor of differentiation 4 (ID4): From development to cancer. Biochim Biophys Acta 1855:92–103
Pellegrino MJ, Parrish DC, Zigmond RE, Habecker BA (2011) Cytokines inhibit norepinephrine transporter expression by decreasing Hand2. Mol Cell Neurosci 46:671–680
Pelster B, Egg M (2015) Multiplicity of hypoxia-inducible transcription factors and their connection to the circadian clock in the zebrafish. Physiol Biochem Zool 88:146–157
Pires N, Dolan L (2010) Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol E 27:862–874
Pruszko M, Zylicz A (2016) [ID proteins in tumour initiation and progression]. Postepy Biochem 62:449–458
Rani A, Greenlaw R, Smith RA, Galustian C (2016) HES1 in immunity and cancer. Cytokine Growth Factor Rev 30:113–117
Riley P, Anson-Cartwright L, Cross JC (1998) The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet 18:271–275
Roschger C, Cabrele C (2017) The Id-protein family in developmental and cancer-associated pathways. Cell Commun Signal 15:7
Rudnicki MA, Le Grand F, McKinnell I, Kuang S (2008) The molecular regulation of muscle stem cell function. Cold Spring Harb Symp Quant Biol 73:323–331
Sailsbery JK, Atchley WR, Dean RA (2012) Phylogenetic analysis and classification of the fungal bHLH domain. Mol Biol E 29:1301–1318
Sato F, Bhawal UK, Yoshimura T, Muragaki Y (2016) DEC1 and DEC2 crosstalk between circadian rhythm and tumor progression. J Cancer 7:153–159
Schoenhard JA, Eren M, Johnson CH, Vaughan DE (2002) Alternative splicing yields novel BMAL2 variants: tissue distribution and functional characterization. Am J Physiol Cell Physiol 283:C103–C114
Schoenhard JA, Smith LH, Painter CA, Eren M, Johnson CH, Vaughan DE (2003) Regulation of the PAI-1 promoter by circadian clock components: differential activation by BMAL1 and BMAL2. J Mol Cell Cardiol 35:473–481
Sherman H, Froy O (2008) Expression of human beta-defensin 1 is regulated via c-Myc and the biological clock. Mol Immunol 45:3163–3167
Shi J, Cao J, Zhou BP (2015) Twist-BRD4 complex: potential drug target for basal-like breast cancer. Curr Pharm Des 21:1256–1261
Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M (2007) Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. Bmc Evol Biol 7:33
Singh K, Dilworth FJ (2013) Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. Febs J 280:3991–4003
Skinner MK, Rawls A, Wilson-Rawls J, Roalson EH (2010) Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation 80:1–8
Song XM, Huang ZN, Duan WK, Ren J, Liu TK, Li Y, Hou XL (2014) Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol Genet Genomics 289:77–91
Stevens JD, Roalson EH, Skinner MK (2008) Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family: genomic approach to cellular differentiation. Differentiation 76:1006–1022
Strazzullo M, Rossetti C, Fusco G, Campanile C, Vecchio D, Campanile G, Perucatti A, Di Meo GP, Filippini F, Eggen A, Ferrara L, D’Esposito M (2010) Genomic characterization and chromosomal mapping of 5 river buffalo skeletal muscle differentiation master genes. Cytogenet Genome Res 128:221–227
Sun H, Fan HJ, Ling HQ (2015) Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16:9
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vostrov AA, Quitschke WW, Vidal F, Schwarzman AL, Goldgaber D (1995) USF binds to the APB alpha sequence in the promoter of the amyloid beta-protein precursor gene. Nucleic Acids Res 23:2734–2741
Wang DP, Li HG, Li YJ, Guo SC, Yang J, Qi DL, Jin C, Zhao XQ (2006) Hypoxia-inducible factor 1alpha cDNA cloning and its mRNA and protein tissue specific expression in domestic yak (Bos grunniens) from Qinghai-Tibetan plateau. Biochem Biophys Res Commun 348:310–319
Wang Y, Chen K, Yao Q, Wang W, Zhi Z (2007) The basic helix-loop-helix transcription factor family in Bombyx mori. Dev Genes E 217:715–723
Wang Y, Chen K, Yao Q, Wang W, Zhu Z (2008a) The basic helix-loop-helix transcription factor family in the honey bee, Apis mellifera. J Insect Sci 8:1–12
Wang Y, Chen KP, Yao Q (2008b) [Progress of studies on bHLH transcription factor families]. Yi Chuan 30:821–830
Wang Y, Chen K, Yao Q, Zheng X, Yang Z (2009) Phylogenetic analysis of zebrafish basic helix-loop-helix transcription factors. J Mol E 68:629–640
Wang Y, Yao Q, Chen KP (2010) [Progress of studies on family members and functions of animal bHLH transcription factors]. Yi Chuan 32:307–330
Wang XH, Wang Y, Zhang DB, Liu AK, Yao Q, Chen KP (2014) A genome-wide identification of basic helix-loop-helix motifs in Pediculus humanus corporis (Phthiraptera: Pediculidae). J Insect Sci 14:195
Wang XH, Wang Y, Liu AK, Liu XT, Zhou Y, Yao Q, Chen KP (2015) Genome-wide identification and analysis of basic helix-loop-helix domains in dog, Canis lupus familiaris. Mol Genet Genomics 290:633–648
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322
Xiong X, Fu M, Lan D, Li J, Zi X, Zhong J (2015) Yak response to high-altitude hypoxic stress by altering mRNA expression and DNA methylation of hypoxia-inducible factors. Anim Biotechnol 26:222–229
Zhang DB, Wang Y, Liu AK, Wang XH, Dang CW, Yao Q, Chen KP (2013) Phylogenetic analyses of vector mosquito basic helix-loop-helix transcription factors. Insect Mol Biol 22:608–621
Zhao Y, Su H, Zhou J, Feng H, Zhang KQ, Yang J (2015) The APSES family proteins in fungi: characterizations, evolution and functions. Fungal Genet Biol 81:271–280
Zheng X, Wang Y, Yao Q, Yang Z, Chen K (2009) A genome-wide survey on basic helix-loop-helix transcription factors in rat and mouse. Mamm Genome 20:236–246
Zhu QQ, Ma C, Wang Q, Song Y, Lv T (2016) The role of TWIST1 in epithelial-mesenchymal transition and cancers. Tumour Biol 37:185–197
Acknowledgements
We are grateful to the anonymous reviewers for their suggestions. This work is supported by National Natural Science Foundation of China (No. 31301965), 2014 Annual Anhui Provincial Project of Outstanding Young Talents Fund in Colleges and Universities (No.[2014]181), and Anhui Provincial Educational Commission Natural Science Foundation (No. KJ2016SD47).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, F., Liu, W. Genome-wide identification, classification, and functional analysis of the basic helix–loop–helix transcription factors in the cattle, Bos Taurus . Mamm Genome 28, 176–197 (2017). https://doi.org/10.1007/s00335-017-9683-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-017-9683-x