Abstract
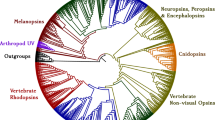
Daphnia pulex has the largest known family of opsins, genes critical for photoreception and vision in animals. This diversity may be functionally redundant, arising from recent processes, or ancient duplications may have been preserved due to distinct functions and independent contributions to fitness. We analyzed opsins in D. pulex and its distant congener Daphnia magna. We identified 48 opsins in the D. pulex genome and 32 in D. magna. We inferred the complement of opsins in the last common ancestor of all Daphnia and evaluated the history of opsin duplication and loss. We further analyzed sequence variation to assess possible functional diversification among Daphnia opsins. Much of the opsin expansion occurred before the D. pulex-D. magna split more than 145 Mya, and both Daphnia lineages preserved most ancient opsins. More recent expansion occurred in pteropsins and long-wavelength visual opsins in both species, particularly D. pulex. Recent duplications were not random: the same ancestral genes duplicated independently in each modern species. Most ancient and some recent duplications involved differentiation at residues known to influence spectral tuning of visual opsins. Arthropsins show evidence of gene conversion between tandemly arrayed paralogs in functionally important domains. Intron–exon gene structure was generally conserved within clades inferred from sequences, although pteropsins showed substantial intron size variation. Overall, our analyses support the hypotheses that diverse opsins are maintained due to diverse functional roles in photoreception and vision, that functional diversification is both ancient and recent, and that multiple evolutionary processes have influenced different types of opsins.






Similar content being viewed by others
References
Arikawa K (2003) Spectral organization of the eye of a butterfly, Papilio. J Comp Physiol A 189:791–800
Barlow HB (1982) What causes trichromacy? A theoretical analysis using comb-filtered spectra. Vis Res 22:635–643
Benzie JAH (2005) The Genus Daphnia (including Daphniopsis): (Anomopoda, Daphniidae). Kenobi Productions, Ghent
Birney E, Clamp M, Durbin R (2004) GeneWise and genomewise. Genome Res 14:988–995
Eriksson BJ, Fredman D, Steiner G, Schmid A (2013) Characterisation and localisation of the opsin protein repertoire in the brain and retinas of a spider and an onychophoran. BMC Evol Biol 13:186
Brandon CS, Dudycha JL (2014) Ecological constraints on sensory systems: compound eye size in Daphnia is reduced by resource limitation. J Comp Physiol A 200:749–758
Brandon CS, James T, Dudycha JL (2015) Selection on incremental variation of eye size in a wild population of Daphnia. J. Evol Biol 28:2112–2118
Briscoe AD (2001) Functional diversification of lepidopteran opsins following gene duplication. Mol Biol Evol 18:2270–2279
Briscoe AD (2002) Letter to the editor homology modeling suggests a functional role for parallel amino acid substitutions. Mol Biol Evol 19:983–986
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al (2009) BLAST+: architecture and applications. BMC Bioinform 10:421
Cohen JH, Richard B, Forward RB (2009) Zooplankton diel vertical migration—a review of proximate control. Oceanogr Mar Biol 47:77–110
Colbourne JK, Hebert PDN (1996) The systematics of North American Daphnia (Crustacea: Anomopoda): a molecular phylogenetic approach. Philos Trans R Soc Lond B 351:349–360
Colbourne JK, Hebert PDN, Taylor DJ (1997) Evolutionary origins of phenotypic diversity in Daphnia. In: Givnish TJ, Sytsma KJ (eds) Molecular evolution and adaptive radiation. Cambridge University Press, Cambridge, pp 163–188
Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH et al (2011) The ecoresponsive genome of Daphnia pulex. Science 331:555–561
De Meester L (1991) An analysis of the phototactic behaviour of Daphnia magna clones and their sexual descendants. Hydrobiologia 225:217–227
De Meester L (1993) Genotype, fish-mediated chemical, and phototactic behavior in Daphnia magna. Ecology 74:1467–1474
Feuda R, Rota-Stabelli O, Oakley TH, Pisani D (2014) The comb jelly opsins and the origins of animal phototransduction. Genome Biol Evol 6(8):1964–1971
Frentiu FD, Bernard GD, Sison-Mangus MP, Van Zandt Brower A, Briscoe AD (2007) Gene duplication is an evolutionary mechanism for expanding spectral diversity in the long-wavelength photopigments of butterflies. Mol Biol Evol 24:2016–2028
Hamza W, Ruggiu D (2000) Swimming behaviour of Daphnia galeata x hyalina as a response to algal substances and to opaque colours. Int Rev Hydrobiol 85:157–166
Hering L, Mayer G (2014) Analysis of the opsin repertoire in the tardigrade Hypsibius dujardini provides insights into the evolution of opsin genes in panarthropoda. Genome Biol Evol 6:2380–2391
Hessen DO, Borgeraas J, Kessler K, Refseth UH (1999) UV-B susceptibility and photoprotection of Arctic Daphnia morphotypes. Polar Res 18:345–352
Hofmann CM, Carleton KL (2009) Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. Integr Comp Biol 49:630–643
Keith N, Tucker AE, Jackson CE, Sung W, Lucas Lledo JI, Schrider DR, Schaack S, Dudycha JL, Shaw J, Lynch M (2016) High mutational rates of large-scale duplication and deletion in Daphnia pulex. Genome Res 26(1):60–69
Kojima D, Mori S, Torii M, Wada A, Morishita R, Fukada Y (2011) UV-sensitive photoreceptor protein OPN5 in humans and mice. PLOS ONE 6(10):e26388
Kotov AA, Taylor DJ (2011) Mesozoic fossils (>145 Mya) suggest the antiquity of the subgenera of Daphnia and their coevolution with chaoborid predators. BMC Evol Biol 11:129
Koyanagi M, Kubokawa K, Tsukamoto H, Shichida Y, Terakita A (2005) Cephalochordate melanopsin: evolutionary linkage between invertebrate visual cells and vertebrate photosensitive retinal ganglion cells. Curr Biol 15:1065–1069
Koyanagi M, Takada E, Nagata T, Tsukamoto H, Terakita A (2013) Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue. Proc Natl Acad Sci USA 110:4998–5003
Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lee YJ, Shah S, Suzuki E, Zars T, O’Day PM, Hyde DR (1994) The Drosophila dgq gene encodes a G α protein that mediates phototransduction. Neuron 13(5):1143–1157
Lewis A, Marcus MA, Ehrenberg B, Crespi H (1978) Experimental evidence for secondary protein-chromophore interactions at the Schiff base linkage in bacteriorhodopsin: molecular mechanism for proton pumping. Proc Natl Acad Sci USA 75:4642–4646
Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A (2003) Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem 278:21655–21662
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Marshall J, Arikawa K (2014) Unconventional colour vision. Curr Biol 24:1150–1154
Marshall J, Carleton KL, Cronin T (2015) Colour vision in marine organisms. Curr Opin Neurobiol 34:86–94
Miner BE, Kerr B (2011) Adaptation to local ultraviolet radiation conditions among neighbouring Daphnia populations. Proc R Soc Lond B 278:1306–1313
Nakane Y, Shimmura T, Abe H, Yoshimura T (2014) Intrinsic photosensitivity of a deep brain photoreceptor. Curr Biol 24:R596–R597
Nathans J (1987) Molecular biology of visual pigments. Annu Rev Neurosci 10:163–194
Oakley TH, Huber DR (2004) Differential expression of duplicated opsin genes in two eyetypes of ostracod crustaceans. J Mol Evol 59:239–249
Ohta T (2010) Gene conversion and evolution of gene families: an overview. Genes (Basel) 1:349–356
Omilian A, Cristescu MEA, Dudycha JL, Lynch M (2006) Ameiotic recombination in asexual lineages of Daphnia. Proc Natl Acad Sci 103:18638–18643. PMC1693715. Erratum: PNAS 104: 2554
Palczewski K, Kumasaka T, Hori T, Behnke CA, Fox BA, Trong I et al (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289:739–745
Porter ML, Cronin TW, McClellan DA, Crandall KA (2007) Molecular characterization of crustacean visual pigments and the evolution of pancrustacean opsins. Mol Biol Evol 24:253–268
Porter ML, Bok MJ, Robinson PR, Cronin TW (2009) Molecular diversity of visual pigments in Stomatopoda (Crustacea). Vis Neurosci 26:255–265
Porter ML, Blasic JR, Bok MJ, Cameron EG, Pringle T, Cronin TW et al (2012) Shedding new light on opsin evolution. Proc R Soc B 279:3–14
Randel N, Bezares-Calderón LA, Gühmann M, Shahidi R, Jékely G (2013) Expression dynamics and protein localization of rhabdomeric opsins in platynereis larvae. Integr Comp Biol 53:7–16
Rennison DJ, Owens GL, Taylor JS (2012) Opsin gene duplication and divergence in ray-finned fish. Mol Phyl Evol 62:986–1008
Rhode SC, Pawlowski M, Tollrian R (2001) The impact of ultraviolet radiation on the vertical distribution of zooplankton of the genus Daphnia. Nature 412:69–72
Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV (2003) remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol 13:1512–1517
Rosenbaum DM, Rasmussen SGF, Kobilka BK (2009) The structure and function of G-protein-coupled receptors. Nature 459:356–363
Sakamoto K, Hisatomi O, Tokunaga F, Eguchi E (1996) Two opsins from the compound eye of the crab Hemigrapsus sanguineus. J Exp Biol 199:441–450
Salcedo E, Zheng L, Phistry M, Bagg EE, Britt SG (2003) Molecular basis for ultraviolet vision in invertebrates. J Neurosci 23:10873–10878
Salcedo E, Farrell DM, Zheng L, Phistry M, Bagg EE, Britt SG (2009) The green-absorbing Drosophila Rh6 visual pigment contains a blue-shifting amino acid substitution that is conserved in vertebrates. J Biol Chem 284:5717–5722
Sawyer S (1989) Statistical tests for detecting gene conversion. Mol Biol Evol 6:526–538
Shichida Y, Matsuyama T (2009) Evolution of opsins and phototransduction. Philos Trans R Soc Lond B 364:2881–2895
Smith FE, Baylor ER (1953) Color responses in the cladocera and their ecological significance. Am Nat 87:49–55
Smith KC, Macagno ER (1990) UV photoreceptors in the compound eye of Daphnia magna (Crustacea, Branchiopoda). A fourth spectral class in single ommatidia. J Comp Physiol A 166:587–606
Speiser DI, Pankey MS, Zaharoff AK, Battelle BA, Bracken-grissom HD, Breinholt JW et al (2014) Using phylogenetically-informed annotation (PIA) to search for light-interacting genes in transcriptomes from non-model organisms. BMC Bioinform 15:1–12
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stearns SC (1975) Light responses of Daphnia pulex. Limnol Oceanogr 20:564–570
Storz UC, Paul RJ (1998) Phototaxis in water fleas (Daphnia magna) is differently influence by visible and UV light. J Comp Phys A 183:709–717
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Terakita A (2005) Protein family review the opsins. Genome Biol 6:1–9
Terakita A, Hariyama T, Tsukahara Y, Katsukura Y, Tashiro H (1993) Interaction of GTP-binding protein Gq with photoactivated rhodopsin in the photoreceptor membranes of crayfish. FEBS Lett 330(2):197–200
Tierney SM, Cooper SJB, Saint KM, Bertozzi T, Hyde J, Humphreys WF et al (2015) Opsin transcripts of predatory diving beetles: a comparison of surface and subterranean photic niches Subject Category: subject Areas: Authors for correspondence: R. Soc Open Sci 2:140386
Velarde RA, Sauer CD, Walden KK, Fahrbach SE, Robertson HM (2005) Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem Mol Biol 35(12):1367–1377
Wakakuwa M, Terakita A, Koyanagi M, Stavenga DG, Shichida Y, Arikawa K (2010) Evolution and mechanism of spectral tuning of blue-absorbing visual pigments in butterflies. PLoS ONE 5:e15015
Yamashita T, Ohuchi H, Tomonari S, Ikeda K, Sakai K, Shichida Y (2010) Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc Natl Acad Sci USA 107:22084–22089
Yan EC, Kazmi MA, De S, Chang BS, Seibert C, Marin EP,… Sakmar TP (2002) Function of extracellular loop 2 in rhodopsin: glutamic acid 181 modulates stability and absorption wavelength of metarhodopsin II. Biochemistry 41(11): 3620–3627
Young S, Taylor VA, Watts E (1984) Visual factors in Daphnia feeding. Limnol Oceanogr 29:1300–1308
Acknowledgements
We appreciate discussion during this project and commentary on earlier drafts of this manuscript from Dan Speiser, Todd Oakley, Sally Woodin, and Axios. The Daphnia Genomics Consortium provided access to the data on which this manuscript is based. CSB was supported by a U.S. Department of Education GAANN Fellowship. D. magna sequence data were produced by The Center for Genomics and Bioinformatics at Indiana University and distributed via wFleaBase in collaboration with the Daphnia Genomics Consortium at http://daphnia.cgb.indiana.edu. D. magna sequencing was supported in part by National Institutes of Health award 5R24GM078274 “Daphnia Functional Genomics Resources.”
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
239_2016_9777_MOESM6_ESM.xlsx
Amino acid alignment of Daphnia opsins by phylogenetic clade (LOPA, LOPB, Arthropsin, etc.) mapped to bovine rhodopsin, and divided by domains following the model presented in Terakita (2005). Amino acid positions conserved across all Daphnia genes with complete data and the bovine rhodopsin are highlighted in yellow (XLSX 383 kb)
Rights and permissions
About this article
Cite this article
Brandon, C.S., Greenwold, M.J. & Dudycha, J.L. Ancient and Recent Duplications Support Functional Diversity of Daphnia Opsins. J Mol Evol 84, 12–28 (2017). https://doi.org/10.1007/s00239-016-9777-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-016-9777-1