Abstract
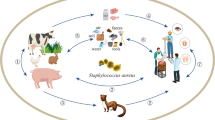
Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) is a relatively recent phenomenon in veterinary medicine. Although in the beginning it was restricted to a single clonal complex (CC), CC398, it has expanded into several clonal complexes, and the diversity of subtypes in the clonal complexes is increasing also. The prevalence of each type is determined somewhat geographically; for instance, the most prevalent clonal complex in Europe is CC398, whereas in Asia, it is CC9. Although few data exist regarding North America, the situation appears to be mixed there. The SCCmec cassettes detected in LA-MRSA are limited mainly to SCCmec IVa and SCCmec V, although non-typeable cassettes and SCCmec type XI, containing mecC, also have been found. The source of the SCCmec in LA-MRSA was discovered to be animals. In searching from which bacteria the SCCmec cassettes in LA-MRSA have been transferred, the most obvious species to consider are the methicillin-resistant non-S. aureus staphylococci (MRNaS). However, very few data are available from those species in animals, and the data that do exist are not detailed enough to determine the origin. Nevertheless, similar cassettes were found in MRNaS, indicating a possible origin that needs to be investigated further.
Similar content being viewed by others
Introduction
Since livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) was detected in pigs in the Netherlands [1], research on MRSA in animals has increased enormously and is far from being complete. Indeed, new findings create new questions regarding how LA-MRSA is evolving. Although it began as a single clonal complex (CC) 398 most of which was sequence type (ST) 398, it is clear now that its diversity is much greater and is rapidly changing over time [2•]. The number of staphylococcal protein A gene (spa) types within CC398 is nowadays increasing [2•]. Furthermore, other S. aureus lineages among animals have acquired methicillin resistance [3]. Moreover, it is obvious that the spread of MRSA in livestock differs geographically, with CC398 being the most prevalent CC in Europe and USA and CC9 predominating in LA-MRSA cases in Asia. Clearly, in Asia LA-MRSA is evolving differently.
Still unanswered is the question, “From where, seemingly all of a sudden, does this methicillin resistance emerge?” Studies have shown that after a host jump of CC398, which was a human-associated clone, to animals, it acquired the staphylococcal cassette chromosome (SCC) mec (SCC) mec [4••]. However, few reports were published regarding the presence of methicillin resistance in staphylococci, and reports on the presence of a methicillin resistance reservoir in staphylococci other than S. aureus were rare. Recently, though, several studies found a high prevalence of methicillin resistance in staphylococci other than S. aureus [5–9, 10••, 11]. Nevertheless, it remains to be determined whether this may be the origin of the SCCmec in livestock-associated S. aureus. Clearly, more research is needed in this field.
Besides its presence in livestock animals, methicillin resistance is being reported more frequently in pet animals (cats and dogs) as well, and this resistance seems to be increasing [12, 13]. The epidemiology in pets, however, appears to be quite different and is limited to some Staphylococcus pseudintermedius and human-derived clones as well as in methicillin-susceptible S. aureus (MSSA) of CC398 [14]. LA-MRSA has been found only in pets residing on farms [13]; therefore, this review will not discuss pet animals.
In this article, we review the current situation regarding LA-MRSA and methicillin resistance in other staphylococci to assess whether the methicillin resistance in S. aureus might originate from other resident staphylococci. After a general description of LA-MRSA, we first provide an overview of the current situation regarding LA-MRSA in different parts in the world and the evolution of this bacterium. Then, we discuss the potential transfer of methicillin resistance from other staphylococci of animal origin to new S. aureus clones.
General Characteristics of LA-MRSA
The first description of LA-MRSA, formerly known as non-typeable (NT) MRSA, included the non-typeability of the strain based on standard pulsed field gel electrophoresis (PFGE) using the restriction enzyme SmaI [1] and was limited to ST398 isolates. Since then, much evolution has occurred, and more sequence types are included now [15, 16]. Although CC398 is still the most common LA-MRSA worldwide, its prevalence differs geographically; in certain regions, other sequence types are involved, such as ST9 in Asia. In addition, the diversity among sequence types is greater in some areas than in others, although the reasons for this remain unclear. In this part, we focus on the most prevalent sequence types found.
CC398
CC398 LA-MRSA is the major clonal complex found in Europe and North America. It occasionally is observed in Asia [17] and also has been detected in Africa [18, 19]. This complex is associated mainly with the colonization of pigs and veal calves [2•, 20, 21••, 22, 23••, 24, 25]. CC398 isolates also have been detected infrequently in poultry [26] and horses [27, 28]. Whole-genome sequencing has shown that this clone originated in humans [4••]; indeed, in humans, CC398 still occurs mainly as MSSA [15, 29•, 30, 31], albeit at a low prevalence. MSSA CC398 also remains present in animals, including pigs [32], dogs [14], bovines [29•], and poultry [29•, 33••]. Currently, CC398 includes 43 sequence types [34], but the major MRSA sequence type colonizing pigs is ST398. Other STs described in pigs are ST541, ST1965, ST1966, ST1967, and ST1968 [16, 35, 36]. However, it has been shown that there is a specific subgroup of human ST398 strains, different from LA-MRSA ST398 [37], that can be readily differentiated by SNP detection and the presence or absence of scn and tet(M) [38]. Research in the Netherlands revealed that all the CC398 strains there typically are LA-MRSA [39]. Nevertheless, the prevalence of MSSA CC398 infections and colonization seems to be increasing, albeit with major geographic differences [40–42].
LA-MRSA CC398 is not considered to be very pathogenic in humans. In animals, however, it has been implicated in bovine mastitis [23••, 43–46] and in infected foot joints in turkeys [33••]. Little information is available regarding pigs because Staphylococcus hyicus is the major pathogenic Staphylococcus species in these animals [47], although S. aureus has been isolated occasionally from lesions in pigs [47]. More recent reports, however, show that LA-MRSA CC398 may infect humans and pigs more often than previously thought [48–50], but this question needs further study.
Typical of this clone is its multiresistance to several classes of antimicrobial agents. In LA-MRSA CC398 strains, typical S. aureus resistance genes have been detected against trimethoprim [dfrA (dfrS1), dfrD, dfrG], tetracycline [tet(K), tet(M), tet(L)], macrolides [msr(A)], lincosamides [lnu(A)], macrolide–lincosamide–streptogramin B [erm(A), erm(B), erm(C)], pleuromutilin–lincosamide–streptogramin A [vga(A)], phenicols (fexA), aminoglycosides (aacA-aphD, aadD, aphA3, spc), and mupirocin (mupA) [51–53]. New resistance genes also have been found frequently in this clone, such as those against aminoglycosides (apmA, spd), trimethoprim (dfrK), macrolide–lincosamide–streptogramin B [erm(T)], and pleuromutilin–lincosamide–streptogramin A [vga(C), vga(E), lsa(E)] [51, 52, 54, 55]. This clone also can easily acquire more rare genes, such as the multiresistance gene cfr (encoding resistance to “PHLOPSA” antibiotics: phenicols, lincosamides, oxazolidines, pleuromutilins, and streptogramins), first found in Staphylococcus sciuri and later in other staphylococci as well as in other genera, including Gram-negative bacteria, which is of great importance given the diversity of antibiotics to which this gene is encoding resistance [56].
The two predominant SCCmec types among CC398 isolates are SCCmec IVa and V; however, IV variants such as SCCmec type IV (2B&5), as well as III and NT types, have been described in bovines [22, 29•] and pigs [2•]. The CC398 isolates firstly identified as carrying SCCmec type III, via the SCCmec PCR typing scheme developed by Zhang et al. [57] corresponded in fact to isolates carrying SCCmec type V [58, 59]. However, LA-MRSA CC398 harboring SCCmec type III was described in bovines [22] and pigs [2•] by researchers using a SCCmec PCR typing scheme developed by Kondo et al. [60].
The ability of MRSA CC398 to acquire foreign DNA may be one of its most dangerous features. It is capable of acquiring virulence genes, and its acquisition of the Panton–Valentine leukocidin (PVL) gene (pvl) has been demonstrated [37, 61, 62]. Regarding other virulence factors, staphylococcal enterotoxins occasionally have been reported in LA-MRSA CC398 in pigs [53, 63–65] and turkeys [33••]. In contrast, genes encoding adhesion factors, proteases, hemolysins, other leukocidins, and superantigen-like proteins have been detected frequently in LA-MRSA CC398 isolates from pigs [2•], poultry [26, 33••], and bovines [22]. CC398 strains of human origin carry genes of the immune evasion cluster (IEC), whereas these genes usually were absent in CC398 isolates from animals [66]. However, IEC genes were detected among isolates recovered from nosocomial infections in horses, as well as in veterinarians [66]. In contrast to most other S. aureus clones, LA-MRSA CC398 shows little host specificity; therefore, it can move easily between hosts and acquire genes. Once this strain becomes more virulent, it may become a very dangerous pathogen for various animal species, including humans.
Although when MRSA CC398 was first discovered there were few sequence types and spa types involved, these seem to be increasing over time, as was shown recently in Belgium for the different spa types involved in colonizing pigs [2•]. This observation implies that the CC398 population is still evolving quite rapidly. However, this variability differs among countries, as shown in the European Food Safety Authority (EFSA) surveillance of 2008 [67]. In addition, in some countries, different spa types may constitute most of the isolates, possibly indicating the local circulation of certain strains. Many experts assume that a regionally high variability is related to the international trade of pigs and that countries with high trade levels have a greater diversity of strains [36, 67, 68].
CC9
CC9 is the major LA-MRSA clone found in Asia, although its prevalence may vary substantially among Asian countries [17]. The first time an ST9 strain was found in Europe was in 2008, in Italy, although the most prevalent ST was the typical European ST398 [3]. Since then, LA-MRSA ST9 isolates have been detected in pigs [48] and poultry [69] from Germany and in retail meat from the UK [70]. However, a recent study shows that LA-MRSA ST9 was present in some European countries before the emergence of CC398 [71].
This sequence type also is found as MSSA in pigs [72]. It was one of the most frequent STs in a study in Minnesota in the USA [73] and was identified as one of the most common MSSA strain in pigs in a study comparing it with a historic collection of MSSA strains [71]. Although the reasons for the differences seen between Asia and the rest of the world remain obscure, it is quite possible that SCCmec was acquired only by Asian strains and not by the European ones. ST9 MRSA has been studied much less extensively than CC398 MRSA; however, in contrast to CC398, it is a typical swine-associated sequence type, although it occasionally may be found infecting humans [74–76]. MRSA ST9 isolates also have been shown to be animal pathogens, as they have been implicated in mastitis in bovines [77].
Similar to CC398 MRSA, the CC9 strains are generally multiresistant, and besides the typical resistance genes, they also carry rare resistance genes against lincosamides [lnu(B)], pleuromutilin–lincosamide–streptogramin A [lsa(E)], and PHLOPSA (cfr) [52, 56]. Several SCCmec types have been found in CC9 strains, including SCCmec III, IV, and V and novel and NT types [17]. This large variety of SCCmec types indicates the high number of acquisitions in these strains, whereas in ST398, the types are mainly IV and V as well as occasional NT SCCmec elements.
Some virulence genes have been found in MRSA ST9, including pig strains with PVL [78]. Studies from several Asian countries and Germany found that more than 90 % of LA-MRSA ST9 isolates carried at least one enterotoxin gene [17]. Moreover, a high detection rate of the toxic shock syndrome toxin (TSST-1) gene (tst), but a negative expression of the TSST-1 phenotype, also was reported in LA-MRSA ST9 isolates [76].
The CC9 isolates show a great variety of spa types with a certain geographic distribution. According to Espinosa-Gongora et al. [71], the first European CC9 isolates were discovered between 1973 and 2009 and carried diverse spa types (t337, t526, t899, t1334, t2498, t3446). The European isolates found recently carry mainly spa types t337 [48], t1430 [48, 69], t1939 [70], and t4794 [3]. CC9 Asian isolates carry diverse spa types, such as t899, t4358, and t337 [17].
CC97
The CC97 clonal complex has undergone quite an evolution. It is a leading cause of bovine mastitis worldwide [43, 77, 79, 80] and is found occasionally in small ruminants, pigs, and humans. It was determined that the human strains originated from a bovine to human host jump believed to have occurred approximately 40 years ago and subsequently acquired methicillin resistance [81]. MSSA CC97 strains are still circulating in humans [82, 83].
This clonal complex has been associated with pigs for quite some time and is found in historic collections dating back to the 1970s as well as in current collections [71]. MRSA ST97 was first discovered in Italy in pigs [3]. Later, CC97 was found in Spain, being the first isolate of a new single locus variant of ST97, namely, ST1379 [84]. In that study, CC97 represented nearly 10 % of the strains, with the remaining being ST398 isolates [84]. Subsequently, CC97 was also found among pigs in Spain [82] and in humans [85, 86].
CC1
MRSA strains of CC1 belong to a very successful human lineage of community-acquired (CA) MRSA. CC1/ST1 is one of the major clones circulating in Italy, where it first was reported in pigs [87, 88] and dairy cows [43], and seems to be spreading to other countries. In 2009, one strain was detected in Denmark [89], and recently a few strains were discovered in veal calves and pigs in Belgium [29•]. In humans, strains of this CC tend to carry the PVL toxin. Fortunately, to date and to our knowledge, PVL-positive MRSA CC1 isolates have not yet been isolated from animals; however, typically present are the IEC genes sak and scn, or the enterotoxine gene sea as in human strains, and they also may carry β-hemolysins, LukF–LukS, LukD–LukE, LukX–LukY γ-hemolysin, enterotoxin H, and superantigen X [87, 90]. The typical strain found in animals belongs to spa type t127 [43, 87–89]. MRSA CC1 also has been implicated in mastitis in cows in Italy [43, 90]. The CC1 strains may carry different SCCmec types, such as SCCmec IV and type 5(5C2).
Other Lineages
On several occasions, human-associated MRSA was found in livestock. A peculiar situation was found in Belgium, where ST239 was found in pigs, bovines, and poultry [2•, 22, 26]. ST239 is a typical human-associated MRSA found in livestock, at low prevalence, only in Belgium. The reason this sequence type was found in livestock only in Belgium is not known, and it would be interesting to follow its evolution. Moreover, the PVL-positive CA-MRSA ST80/t044 SCCmec IV European clone was detected recently among pigs in Belgium [2•].
Similarly, strains of ST5 (belonging to CC5) commonly associated with human infections have been isolated from pork and pigs in the USA [91–93]. Unlike ST293, the prevalence of this CC frequently is higher than that of CC398. CC5 strains also have been detected in Canada [94]. A striking observation regarding CC5 is that strains of this lineage, especially ST5, have had host jumps to poultry, in which it frequently is implicated in disease [95]. In fact, broiler chicken and turkey ST5 isolates with avian-niche-specific genes of the φAvβ prophage have been described [4••, 33••]. The significance of ST5 in pigs and on pork remains unclear; however, besides human contamination, which is unlikely despite the high prevalence in some studies, it also may be a new emerging LA-MRSA clone in the USA, as it was shown recently that ST5 is one of the three most frequent MSSA clones found in pigs in Minnesota [73]. Further studies on ST5 are warranted to determine its origin and relationship to human or poultry strains. The evolution of these strains also should be followed closely given their close relationship to highly pathogenic and hospital (health care)-acquired (HA) human MRSA strains.
Occasionally, sequence types are found that are associated not only with humans. Sequence types specifically associated with a certain livestock host are being reported more frequently, suggesting the spread of SCCmec among other animal S. aureus clones. MRSA CC30 was found recently in pigs from Denmark [89], and isolates from several CCs (CC8, CC9, CC20, CC30, CC45, CC479, CC522, and CC705) have been described among bovine and pig isolates [22, 29•].
SCC mec Type XI, mecC-carrying MRSA From Animals
SCCmec type XI, containing a new mecC gene (formerly named mecA LGA251), was first described from a strain originating from mastitis in cows and from humans in the UK and Denmark [96]. It was discovered because it was negative on mecA PCR but phenotypically resistant. Although this SCCmec type is associated mainly with CC130, it also has been found in CC1943 and CC425 as well as in many other CCs, including CC599 and CC59 [97, 98, 99].
Subsequently, other laboratories started testing for mecC on their mecA-negative strains showing phenotypic resistance. All these studies indicate only a low prevalence; however, these mecC-positive strains appear to be widespread, at least in Europe, and seem to be present in different animal species, including humans, as well as in the environment [99, 100]. The livestock animal species involved are mainly dairy cattle, and these strains have not been recovered from pigs or poultry so far. Other animals involved include sea mammals, pet animals, wildlife (birds and mammals), and zoo animals [97–99, 101–106]. Of note, the ST130 strains carrying SCCmec type XI are likely to be zoonotic and to cause infections in animals [98, 107]. Typically, they carry a diverse array of virulence factors, such as hemolysins, immune evasion factors, enterotoxins, and/or TSST-1 [98].
To our knowledge, mecC has never been found outside Europe, although that may be because few studies have looked for it. Some studies in the USA have looked for but failed to detect it (Tara Smith, personal communication). Nevertheless, there are indications that the mecC strains have a limited geographic spread, as was demonstrated for the strains isolated in France [107].
The SCCmec type XI is divergent in the mec region (it carries a β-lactamase blaZ gene together with the mec operon genes) and in other parts of the cassette compared to other SCCmec types [96]. This divergence indicates that it had an independent evolution, although it is not yet known how it evolved. It also indicates that although the earliest strain found was from 1975, this type may have been circulating for an even longer period [97, 98]. This appears similar to what is observed for mecA-carrying SCCmec elements in coagulase-negative staphylococci; however, for mecC of course fewer strains are available to date, and the diversity is still difficult to assess. The gene mecC has been detected in Staphylococcus xylosus, S. sciuri, and Staphylococcus stepanovicii [108–110]. The mecC allotypes in the S. sciuri and S. xylosus strains are different and are integrated in different SCCmec structures, demonstrating the potential diversity in SCCmec mecC-carrying elements in staphylococci other than S. aureus. However, to our knowledge, no real surveillance has been performed on mecC in staphylococci other than S. aureus. In addition, the prevalence of coagulase-negative strains appears low, although this observation is based merely on the fact that for most strains tested to date (of which there are few) mecA rather than mecC was present [see “Methicillin Resistance in Staphylococci Other Than S. aureus (MRNaS) From Animals and the Possibility of Their Creating New LA-MRSA Clones” section]. It should be noted also that the S. sciuri strain carries both a mecA and a mecC gene, and likewise it may be that more mecA-positive strains carry an additional mecC. Further studies are necessary to determine the role of mecC-carrying elements in methicillin resistance in staphylococci other than S. aureus.
Evolution of LA-MRSA in Europe
LA-MRSA was first detected in the Netherlands [1] and France [111] around the middle of the first decade of the twenty-first century. Before that, reports of methicillin resistance in S. aureus appeared occasionally, the first one on a case of MRSA of human origin isolated from milking cows [112]. However, most of these reports lacked typing data and were based only on phenotypic detection; therefore, they should be interpreted with care. The first confirmed case of LA-MRSA in 2004 in the Netherlands was detected by accident, in a young girl who was colonized by MRSA and could not be decolonized [1, 113]. Upon further investigation into the causes, it was discovered that the source of this MRSA was the pigs on the farm where the girl and her family lived. After this lineage was found to be highly prevalent, many European countries became concerned regarding their own situation. Neighboring countries with similar animal-rearing practices began their own surveillance and found a high prevalence of MRSA in their pigs also [24, 114]. These findings ultimately led to a multinational European surveillance system to detect MRSA in pigs. Unfortunately, the sampling used by the system consisted of dust samples, which later were shown to have a low sensitivity. Moreover, the isolation method used (double-selective enrichment) was shown to be suboptimal in low-prevalence populations, such as poultry [26]. These limitations resulted in a lower estimated prevalence, which also is exemplified by the lower prevalence in Belgium during that study. Whereas the first surveillance showed a prevalence of approximately 60 % [20], the EFSA study identified a prevalence of only about 40 % [67], and a subsequent surveillance found a prevalence greater than 60 % [2•]. These results demonstrate that uniform sampling and isolation are of the utmost importance in comparing data.
Prevalence rates among European countries differ substantially. In some countries, no MRSA was found, whereas in others, the prevalence was high and there was a whole in-between group [68]. As stated earlier, the sensitivity of this study was quite low, and the research did not allow sampling from, for example, pigs raised outdoors, which is still done frequently in some countries. Although dust sampling from outdoor-reared pigs is rather irrelevant, the reasons for this high variability remain quite obscure; there certainly is an indication that international trade increased the spread of LA-MRSA CC398.
The CC398 strains initially were named NT-MRSA, because their genomic DNA could not be digested with the SmaI endonuclease, the enzyme used in the PFGE method for S. aureus. This was a result of the action of C5-cytosine methyltransferase, which modifies the consensus sequence recognized by SmaI [115, 116]. Although several enzymes were used as an alternative [117], their profiles were not comparable with those generated by SmaI in non-CC398 isolates. To overcome this problem, the use of Cfr9I, a neoschizomer (an enzyme that cuts within the same recognition sequence) of SmaI, was proposed [116], and this enzyme was used successfully for PFGE typing of CC398 isolates [59]. Although the use of Cfr9I allowed direct comparison with SmaI–PFGE profiles, differentiation of LA-MRSA CC398 was based mainly on SCCmec and spa typing. Initially, CC398 SCCmec was thought to be type III; however, later it was proven that it was mainly types IV and V, and the isolates typed as SCCmec III corresponded to type V [58, 59]. To date, little variation has been seen in the prevalence of CC398 strains; however, with increased use of zinc oxide in some European countries, SCCmec type V (5C2&5) may increase. Indeed, after the discovery of the zinc oxide resistance gene (czrC) in MRSA CC398 [118] and its location on SCCmec type V [119–121], a randomized controlled trial and an epidemiologic study showed that zinc oxide selects for MRSA CC398 SCCmec type V in vivo [122, 124]. The possible co-selection of MRSA through zinc oxide application in pigs is supported further by pig experiments showing that treatment with zinc oxide alone and in combination with tetracycline influences the MRSA load in the nasal cavity [124]. Besides SCCmec types IV and V, other types have been described rarely, and these are mainly NT SCCmec types [2•, 22, 29•]. However, SCCmec also undergoes an evolution, as exemplified by the fact that there are MSSA CC398 strains with an SCC remnant V(5C2&5) from which the mec gene complex has been deleted. Strikingly, this element still carries the zinc and tetracycline resistance genes [29•].
The history of MRSA CC398 is young, with scientific investigation starting a little more than 10 years ago. However, it must have been circulating in pigs earlier, given its high prevalence in some countries, although it is difficult to estimate how long it has been circulating. In their recent study, Espinosa-Gongora et al. [71] found that ST398 was absent in collections of pig isolates recovered between 1973 and 2003. The authors suggested that CC398 was either absent or present at low frequencies in pigs in the past and confirmed the current theory that S. aureus ST398 did not originate in pigs. The period during which CC398 has been circulating might be rather short given the high transmission ratios [21] and the intense trade of pigs throughout Europe [36, 67, 68]. Thus, it is supposed that the evolution of this clone in animals began quite recently. Indeed, there are some indications that this population is in an early stage of evolution because the diversity of spa types found in specific geographic regions is still increasing [2•]. Moreover, recent papers [29•] also indicate other lineages arising within the pig population, highlighting the continuous evolution of MRSA dynamics in this species and the need for continuous surveillance so that if new and perhaps more dangerous clones arise, early intervention strategies may be executed. Because the main route by which these new MRSA types spread is trade [68], it is clear that new trade policies must be developed to prevent the spread. For countries in which MRSA is absent, it is especially important to import only MRSA-free pigs. However, there currently is no legal basis for preventing the entry of MRSA pigs or other animals so that these countries can protect their livestock from becoming colonized with MRSA.
At the moment, it is difficult to predict how MRSA will evolve. We know that in countries where it is highly prevalent, the prevalence will not decrease. Positive farms will remain positive, and there is no indication they will become negative [2•]. In some countries, negative farms will become difficult to find; moreover, the problem will affect not only pigs, but bovines as well [22, 23••].
In bovines, controlling cases of S. aureus mastitis, as well as those caused by MRSA, has proven profitable, although some pitfalls must be taken into account. First, eliminating S. aureus mastitis is not easy; it involves the culling of animals testing positive. Second, LA-MRSA also is carried in the nose [22], which may complicate its elimination. Unfortunately, the transmission ratio of LA-MRSA in adult cattle is unknown; therefore, it is impossible to estimate the success of eradication if other animals, colonized only in the nose, need to be culled. The estimated prevalence of nose carriage is approximately 10 %, and it is unknown whether these animals also carry the bacterium in their udders [22, 23••].
Evolution of LA-MRSA in North America
Studies of LA-MRSA in North America are several years behind those in Europe. They also have been hampered by the increased difficulty of obtaining on-farm samples because of reduced government regulation of farming compared to many European countries. Nevertheless, several studies have been carried out in cooperation with farmers and farming groups to investigate the epidemiology of LA-MRSA in North America. To date, however, no studies examining this issue have been published in Mexico, and those from the USA and Canada are limited and represent the findings from only a handful of states and provinces.
Although ST398 is identified commonly in pigs and farming environments in North America [73, 92–94, 125–133], there appears to be a greater diversity of molecular types found on North American farms compared with those in Europe, where ST398 remains the dominant or sole strain of MRSA identified. One study in Michigan found no ST398 present in the pigs sampled [134], whereas the bulk of studies have found a mix of “human” types (such as ST8 and ST5) in conjunction with ST398.
Typing of SCCmec has been carried out in only a few North American studies. Smith et al. [127] typed all human strains and 15 representative swine isolates, all ST398; all were found to be type V. Molla et al. [93] tested all MRSA isolates (n = 99), among which they identified type V (16 %), type II (7 %), and type IV (5.1 %); they reported 4 % as NT. The authors also noted that “although the majority of the MRSA isolates (67 %) had identifiable ccr gene and mec gene complexes, the combinations we found did not match the currently reported types, suggesting that, like in Europe, a new SCCmec type(s) might be circulating in the porcine isolates.” Like the diversity of spa types and STs found in North American pigs, that of SCCmec types also does not lend itself easily to comparison elsewhere.
Evolution of LA-MRSA in Asia
The situation in Asia is very different from the one in Europe and USA. In most Asian countries, CC9 predominates among livestock, in contrast to the widespread CC398-MRSA in Europe and North America [17]. MRSA ST9 and variants have been isolated from pigs in China, Hong Kong, Malaysia, Thailand, Japan, and South Korea [17], but other MRSA lineages, such as ST22, ST221, and ST398, occasionally have been reported among pigs from Asian countries [17].
In Asia, the overall carriage rate of MRSA in pigs is generally low. The prevalence of CC9, the most prevalent MRSA lineage among pigs, varies widely: 1.4 % in Malaysia [135], 10 % in Thailand [136], 4 to 13.9 % in Taiwan [137, 138], 11.4 to 14.7 % in China [139, 140], and 21.3 % in Hong Kong [141]. However, a recent study found a high nasal rate (40 %) of MRSA CC9 among pigs in Thailand [142]. MRSA CC9 isolates also have been detected in dust samples from pig farms in China [143] and at a low frequency in chickens and meat (chicken and beef) samples from Hong Kong [141, 144]. Similarly, MRSA CC9 among pig farmers in Asian countries also appears low (5–19 %) [135, 138, 139].
Although most LA-MRSA CC9 isolates in Asia are typed as ST9, single-locus variants (such as ST1376) also have been found [143]. LA-MRSA ST9 isolates carry different types of SCCmec depending on the country: types IV and V in Taiwan [78]; types III and IVb in China [139, 140]; type IV, IVb, or V in Hong Kong [141, 144, 145]; type V in Malaysia [135]; and type IX in Thailand [142, 146]. LA-MRSA CC9 isolates with NT SCCmec cassettes also have been described in Taiwan [78, 137, 138], and recently a novel SCCmec type with ccrAB type 1 and mec class C was found in ST9-t337 from Thailand [136]. Moreover, distribution of the different spa types also is country related: t899 predominates in China [139, 140] and Hong Hong [141, 144], t4358 in Malaysia [135], and t337 in Thailand [76, 136]; however, other related spa types (such as t2922) have been found in LA-MRSA CC9 [140].
A recent study proved that the Taiwan clone of LA-MRSA ST9 and human clinical ST9-MRSA belong to a novel staphylocoagulase (SC) XIc subtype [76]. In this study, ST9 MRSA isolates of human and swine origin showed a highly homogeneous virulence genotype and genomic profiles. The authors suggested the existence of a recent common ancestor and a cross-species transmission of the emerging ST9-SCXIc MRSA between swine and humans [76].
LA-MRSA CC398, including ST398-t034 and ST541-t034, initially were found at a low carriage rate (2.6 %) in commercial pigs from South Korea [16]. However, these isolates may have an American or European origin, as breeding pigs were imported from the USA, Canada, and Denmark [16]. In this study, few isolates (prevalence of 0.6 %) were typed as ST72-SCCmec IVa PVL-negative, which corresponded to the CA-MRSA lineage more prevalent in that country [16]. MSSA CC398 isolates also have been reported occasionally in pigs from Japan [72], as well as at a high carriage rate (16.8 %) in pigs from China [140]. In a research hospital in Singapore, MRSA ST398 isolates also were found in experimental pigs obtained from Indonesia [147].
Regarding other lineages, LA-MRSA ST221-t002 was isolated from a swine nasal sample (0.9 %) in Japan [148], and an ST22-SCCmec IV isolate also was isolated in the experimental pigs of the Singapore research hospital [147].
LA-MRSA in Other Parts of the World
Little is known about the prevalence of LA-MRSA in developing countries. Although few studies have been performed on the epidemiology of LA-MRSA in Africa [18, 149–152], several lineages have been detected, such as the human-associated ST5-SCCmec IV and ST88-SCCmec IV in pigs from Senegal [151] and ST153-SCCmec NT from healthy sheep in Tunisia [152]. Moreover, one veterinarian from Tunisia carried an ST80-SCCmec IVc isolate [153], but interestingly, human-associated S. aureus lineages have been described in chimpanzees, possibly as the result of humanosis [18].
Methicillin Resistance in Staphylococci Other Than S. aureus (MRNaS) From Animals and the Possibility of Their Creating New LA-MRSA Clones
To date, methicillin resistance has been detected in most staphylococcal species, demonstrating that SCCmec perhaps is more mobile than suspected, especially considering that most S. aureus (the most-studied species) clones are not very competent in taking up foreign DNA. It has been shown that specific restriction modification systems in S. aureus are capable of blocking horizontal gene transfer [154]. These restriction modification systems are lineage specific [155], which is exemplified by the fact that frequently in ecosystems, only a few methicillin-resistant clones are circulating whereas the diversity of the methicillin-susceptible clones is much greater. A perfect example of this is the limited number of LA-MRSA clones compared with MSSA clones infecting animals and the introduction of methicillin resistance in animal-associated S. aureus, which occurred only recently, as discussed earlier.
Clearly, the SCCmec types found in MRNaS are similar to those in S. aureus, although the variability in MRNaS appears much greater, including the presence of NT cassettes. It should be noted that few data exist for most of the MRNAS, making it difficult to assess the variability of SCCmec. Studies also have been performed in which the SCCmec cassettes have been characterized further [5–9, 10••, 11, 46, 156, 157].
However, these analyses are not detailed enough to demonstrate the transfer of SCCmec. As shown [158], there is much more variation in the sequences, and ideally the SCCmec types should be subdivided based on differences in sequences. This task has become feasible with new-generation sequencing; therefore, it no longer will be a great challenge to develop a more detailed subtyping scheme of SCCmec types to enable researchers to follow their movements, plasticity, and epidemiology.
Clearly, the prevalence of the different SCCmec types in MRNaS also differs from that of S. aureus. This difference simply may be a result of the possibility that—based on epidemiologic data, as presented earlier—staphylococci other than S. aureus might acquire SCCmec more easily; however, this needs further confirmation.
Conclusions
It is clear that the story of LA-MRSA is not over. New challenges may arise with the diversification of LA-MRSA, through which new spa types might arise, and new sequence types might acquire methicillin resistance from the large reservoir that exists in other staphylococcal species, although the latter requires more definitive characterization.
The prevalence of LA-MRSA is low in some countries and high in others. Some countries, such as Norway, are trying to become negative for LA-MRSA; however, this appears very challenging [159]. Based on our current knowledge of the epidemiology of methicillin resistance in staphylococci, it is difficult to forecast what the future holds. Certainly, LA-MRSA is a recent event, and it requires more research on methicillin resistance in all staphylococcal species in order to estimate its risk for both animal and human health.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11:1965–66.
Peeters LE, Argudín MA, Azadikhah S, Butaye P. Antimicrobial resistance and population structure of Staphylococcus aureus recovered from pigs farms. Vet Microbiol. 2015;180:151–6.Large diversity of spa types among CC398, and description of the community-acquired (CA-)MRSA ST80-IV European clone among pigs.
Battisti A, Franco A, Merialdi G, et al. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet Microbiol. 2010;142:361–6.
Price LB, Stegger M, Hasman H, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio. 2012;3:e00305–11. Evidence of CC398 human to animal host jump.
Argudín MA, Vanderhaeghen W, Vandendriessche S, et al. Antimicrobial resistance and population structure of Staphylococcus epidermidis recovered from animals and humans. Vet Microbiol. 2015;178:105–13.
Argudín MA, Vanderhaeghen W, Butaye P. Diversity of antimicrobial resistance and virulence genes in methicillin-resistant non-Staphylococcus aureus staphylococci from veal calves. Res Vet Sci. 2015;99:10–6.
Argudín MA, Vanderhaeghen W, Butaye P. Antimicrobial resistance and population structure of Staphylococcus epidermidis recovered from pig farms in Belgium. Vet J. 2015;203:302–8.
Nemeghaire S, Vanderhaeghen W, Argudín MA, Haesebrouck F, Butaye P. Characterization of methicillin-resistant Staphylococcus sciuri isolates from industrially raised pigs, cattle and broiler chickens. J Antimicrob Chemother. 2014;69:2928–34.
Nemeghaire S, Argudín MA, Haesebrouck F, Butaye P. Molecular epidemiology of methicillin-resistant Staphylococcus sciuri in healthy chickens. Vet Microbiol. 2014;171:357–63.
Vanderhaeghen W, Vandendriessche S, Crombé F, et al. Species and staphylococcal cassette chromosome mec (SCCmec) diversity among methicillin-resistant non-Staphylococcus aureus staphylococci isolated from pigs. Vet Microbiol. 2012;158:123–8. First general description of the diversity of SCC mec in staphylococci of animals.
Vanderhaeghen W, Vandendriessche S, Crombé F, et al. Characterization of methicillin-resistant non-Staphylococcus aureus staphylococci carriage isolates from different bovine populations. J Antimicrob Chemother. 2013;68:300–7.
Vanderhaeghen W, Van de Velde E, Crombé F, et al. Screening for methicillin-resistant staphylococci in dogs admitted to a veterinary teaching hospital. Res Vet Sci. 2012;93:133–6.
Pletinckx LJ, Verhegghe M, Crombé F, et al. Evidence of possible methicillin-resistant Staphylococcus aureus ST398 spread between pigs and other animals and people residing on the same farm. Prev Vet Med. 2013;109:293–303.
Gómez-Sanz E, Torres C, Benito D, Lozano C, Zarazaga M. Animal and human Staphylococcus aureus associated clonal lineages and high rate of Staphylococcus pseudintermedius novel lineages in Spanish kennel dogs: predominance of S. aureus ST398. Vet Microbiol. 2013;166:580–9.
Lozano C, Rezusta A, Gómez P, et al. High prevalence of spa types associated with the clonal lineage CC398 among tetracycline-resistant methicillin-resistant Staphylococcus aureus strains in a Spanish hospital. J Antimicrob Chemother. 2012;67:330–4.
Lim SK, Nam HM, Jang GC, Lee HS, Jung SC, Kwak HS. The first detection of methicillin-resistant Staphylococcus aureus ST398 in pigs in Korea. Vet Micro-biol. 2012;155:88–92.
Chuang YY, Huang YC. Livestock-associated meticillin-resistant Staphylococcus aureus in Asia: an emerging issue? Int J Antimicrob Agents. 2015;45:334–40.
Abdulgader SM, Shittu AO, Nicol MP, Kaba M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol. 2015;6:348.
Chairat S, Gharsa H, Lozano C, et al. Characterization of Staphylococcus aureus from raw meat samples in Tunisia: detection of clonal lineage ST398 from the African continent. Foodborne Pathog Dis. 2015;12:686–92.
Crombé F, Willems G, Dispas M, et al. Prevalence and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus among pigs in Belgium. Microb Drug Resist. 2012;18:125–31.
Crombé F, Argudín MA, Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P. Transmission dynamics of methicillin-resistant Staphylococcus aureus in pigs. Front Microbiol. 2013;4:57. First revision of the high transmission ration in pigs, showing that LA-MRSA has features to become endemic.
Nemeghaire S, Argudín MA, Haesebrouck F, Butaye P. Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureus nasal carriage isolates from bovines. BMC Vet Res. 2014;10:153.
Vanderhaeghen W, Cerpentier T, Adriaensen C, Vicca J, Hermans K, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet Microbiol. 2010;144:166–71. First description of LA-MRSA implicated in bovine mastitis.
Verhegghe M, Pletinckx LJ, Crombé F, et al. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 in pig farms and multispecies farms. Zoonoses Public Health. 2013;60:366–74.
Verhegghe M, Crombé F, Pletinckx LJ, et al. Genetic diversity of livestock-associated MRSA isolates obtained from piglets from farrowing until slaughter age on four farrow-to-finish farms. Vet Res. 2014;45:89.
Nemeghaire S, Roelandt S, Argudín MA, Haesebrouck F, Butaye P. Characterization of methicillin-resistant Staphylococcus aureus from healthy carrier chickens. Avian Pathol. 2013;42:342–6.
Van den Eede A, Martens A, Feryn I, et al. Low MRSA prevalence in horses at farm level. BMC Vet Res. 2012;8:213.
Van den Eede A, Hermans K, Van den Abeele A, et al. The nasal vestibulum is the optimal sampling site for MRSA screening in hospitalised horses. Vet J. 2013;197:415–9.
Vandendriessche S, Vanderhaeghen W, Larsen J, et al. High genetic diversity of methicillin-susceptible Staphylococcus aureus (MSSA) from humans and animals on livestock farms and presence of SCC mec remnant DNA in MSSA CC398. J Antimicrob Chemother. 2014;69:355–62. Evidence of SCC mec remnants in MSSA CC398.
Benito D, Aspiroz C, Gilaberte Y, Sanmartín R, Hernández-Martin Á, Alonso M, Gómez P, Lozano C, Torres C. Genetic lineages and antimicrobial resistance genotypes in Staphylococcus aureus from children with atopic dermatitis: detection of clonal complexes CC1, CC97 and CC398. J Chemother. 2015; 1:1973947815Y0000000044.
Uhlemann AC, Dumortier C, Hafer C, et al. Molecular characterization of Staphylococcus aureus from outpatients in the Caribbean reveals the presence of pandemic clones. Eur J Clin Microbiol Infect Dis. 2012;31:505–11.
Hasman H, Moodley A, Guardabassi L, Stegger M, Skov RL, Aarestrup FM. Spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet Microbiol. 2010;141:326–31.
Argudín MA, Cariou N, Salandre O, Le Guennec J, Nemeghaire S, Butaye P. Genotyping and antimicrobial resistance of Staphylococcus aureus isolates from diseased turkeys. Avian Pathol. 2013;42:572–80. First description of LA-MRSA CC398 involved in pathological processes in turkeys.
Stegger M, Aziz M, Chroboczek T, et al. Genome analysis of Staphylococcus aureus ST291, a double locus variant of ST398, reveals a distinct genetic lineage. PLoS ONE. 2013;8:e63008.
Porrero MC, Wassenaar TM, Gómez-Barrero S, et al. Detection of methicillin-resistant Staphylococcus aureus in Iberian pigs. Lett Appl Microbiol. 2012;54:280–5.
Anonymous. European Food Safety Authority. Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008, Part A: MRSA prevalence estimates; on request from the European Commission. EFSA J. 2009;7:1376.
Stegger M, Lindsay JA, Sørum M, Gould KA, Skov R. Genetic diversity in CC398 methicillin-resistant Staphylococcus aureus isolates of different geographical origin. Clin Microbiol Infect. 2010;16:1017–9.
Stegger M, Liu CM, Larsen J, et al. Rapid differentiation between livestock-associated and livestock-independent Staphylococcus aureus CC398 clades. PLoS ONE. 2013;8:e79645.
Lekkerkerk WS, van Wamel WJ, Snijders SV, et al. What is the origin of livestock-associated methicillin-resistant Staphylococcus aureus clonal complex 398 isolates from humans without livestock contact? An epidemiological and genetic analysis. J Clin Microbiol. 2015;53:1836–41.
Valour F, Tasse J, Trouillet-Assant S, et al. Lyon Bone and Joint Infection study group. Methicillin-susceptible Staphylococcus aureus clonal complex 398: high prevalence and geographical heterogeneity in bone and joint infection and nasal carriage. Clin Microbiol Infect. 2014;20:772–5.
Sarrou S, Liakopoulos A, Chasioti M, et al. Dissemination of Methicillin-Susceptible CC398 Staphylococcus aureus strains in a rural Greek area. PLoS ONE. 2015;10:e0122761.
Cuny C, Layer F, Köck R, Werner G, Witte W. Methicillin susceptible Staphylococcus aureus (MSSA) of clonal complex CC398, t571 from infections in humans are still rare in Germany. PLoS ONE. 2013;8:e83165.
Luini M, Cremonesi P, Magro G, et al. Methicillin-resistant Staphylococcus aureus (MRSA) is associated with low within-herd prevalence of intra-mammary infections in dairy cows: genotyping of isolates. Vet Microbiol. 2015;178:270–4.
Spohr M, Rau J, Friedrich A, et al. Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health. 2011;58:252–61.
Tavakol M, Riekerink RG, Sampimon OC, van Wamel WJ, van Belkum A, Lam TJ. Bovine-associated MRSA ST398 in the Netherlands. Acta Vet Scand. 2012;1(54):28.
Silva NC, Guimarães FF, Manzi MP, et al. Methicillin-resistant Staphylococcus aureus of lineage ST398 as cause of mastitis in cows. Lett Appl Microbiol. 2014;59:665–9.
Foster AP. Staphylococcal skin disease in livestock. Vet Dermatol. 2012;23:342–51.
Meemken D, Blaha T, Tegeler R, et al. Livestock associated methicillin-resistant Staphylococcus aureus (LaMRSA) isolated from lesions of pigs at necropsy in northwest Germany between 2004 and 2007. Zoonoses Public Health. 2010;57:e143–8.
van der Wolf PJ, Rothkamp A, Junker K, de Neeling AJ. Staphylococcus aureus (MSSA) and MRSA (CC398) isolated from post-mortem samples from pigs. Vet Microbiol. 2012;158:136–41.
Tristan A, Rasigade JP, Ruizendaal E, Laurent F, Bes M, Meugnier H, Lina G, Etienne J, Celard M, Tattevin P, Monecke S, Le Moing V, Vandenesch F; French AEPEI study Group on Infective Endocarditis. Rise of CC398 lineage of Staphylococcus aureus among Infective endocarditis isolates revealed by two consecutive population-based studies in France. PLoS One. 2012;7:51172.
Kadlec K, Fessler AT, Hauschild T, Schwarz S. Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2012;18:745–55.
Wendlandt S, Feßler AT, Monecke S, Ehricht R, Schwarz S, Kadlec K. The diversity of antimicrobial resistance genes among staphylococci of animal origin. Int J Med Microbiol. 2013;303:338–49.
Jamrozy DM, Fielder MD, Butaye P, Coldham NG. Comparative genotypic and phenotypic characterisation of methicillin-resistant Staphylococcus aureus ST398 isolated from animals and humans. PLoS ONE. 2012;7:e40458.
Wendlandt S, Feßler AT, Kadlec K, van Duijkeren E, Schwarz S. Identification of the novel spectinomycin resistance gene spd in a different plasmid background among methicillin-resistant Staphylococcus aureus CC398 and methicillin-susceptible S. aureus ST433. J Antimicrob Chemother. 2014;69:2000–3.
Jamrozy DM, Coldham NG, Butaye P, Fielder MD. Identification of a novel plasmid-associated spectinomycin adenyltransferase gene spd in methicillin-resistant Staphylococcus aureus ST398 isolated from animal and human sources. J Antimicrob Chemother. 2014;69:1193–6.
Kehrenberg C, Cuny C, Strommenger B, Schwarz S, Witte W. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob Agents Chemother. 2009;53:779–81.
Zhang K, McClure JA, Sameer E, Louie T, Conly JM. Novel multiple PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to IV in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–33.
Jansen MD, Box ATA, Fluit AC. SCCmec-typing in methicillin-resistant Staphylococcus aureus strains of animal origin. Emerg Infect Dis. 2009;15:136–7.
Argudín MA, Fetsch A, Tenhagen BA, et al. High heterogeneity within methicillin-resistant Staphylococcus aureus ST398 isolates, defined by Cfr9I macrorestriction-pulsed- field gel electrophoresis profiles and spa and SCCmec types. Appl Environ Microbiol. 2010;76:652–8.
Kondo Y, Ito T, Ma XX, et al. Combination of multiplex PCRs for Staphylococcal cassette chromosome mec type assignement: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–74.
van Belkum A. Dutch Working Party on Surveillance and Research of MRSA-SOM. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg Infect Dis. 2008;14:479–83.
Yu F, Chen Z, Liu C, et al. Prevalence of Staphylococcus aureus carrying Panton-Valentine leukocidin genes among isolates from hospitalised patients in China. Clin Microbiol Infect. 2008;14:381–4.
Kadlec K, Ehricht R, Monecke S, et al. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J Antimicrob Chemother. 2009;64:1156–64.
Laurent F, Jouy E, Granier S, Bes M, Ruimy R, Felix B, Le Roux A, Sanders P, Chauvin C, Etienne J, Brisabois A.. Molecular characterization of antimicrobial resistance genes and virulence genes by using microarrays in representative ST398 MRSA isolates from pigs in France. Proceedings of the ASM-ESCMID Conference on Methicillin- resistant Staphylococci in Animals, 2009: Veterinary and Public Health Implications (p. 25). London, UK.
Argudín MA, Tenhagen BA, Fetsch A, et al. Guerra B Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Appl Environ Microbiol. 2011;77:3052–60.
Cuny C, Abdelbary M, Layer F, Werner G, Witte W. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet Microbiol. 2015;177:219–23.
Anonymous. European Food Safety Authority. Scientific opinion of the panel on biological hazards on a request from the European Commission on Assessment of the Public Health significance of methicillin resistant Staphylococcus aureus (MRSA) in animals and foods. EFSA J. 2009;1–73.
Anonymous. European Food Safety Authority. Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008—Part B: factors associated with MRSA contamination of holdings. EFSA J. 2010;8:1597.
Fessler AT, Kadlec K, Hassel M, et al. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl Environ Microbiol. 2011;77:7151–7.
Dhup V, Kearns AM, Pichon B, Foster HA. First report of identification of livestock-associated MRSA ST9 in retail meat in England. Epidemiol Infect. 2015;143:2989–92.
Espinosa-Gongora C, Moodley A, Lipinska U, et al. Phenotypes and genotypes of old and contemporary porcine strains indicate a temporal change in the S . aureus population structure in pigs. PLoS One. 2014;9, e101988.
Asai T, Hiki M, Baba K, Usui M, Ishihara K, Tamura Y. Presence of Staphylococcus aureus ST398 and ST9 in Swine in Japan. Jpn J Infect Dis. 2012;65:551–2.
Linhares LL, Yang M, Sreevatsan S, Munoz-Zanzi CA, Torremorell M, Davies PR. The effect of anatomic site and age on detection of Staphylococcus aureus in pigs. J Vet Diagn Invest. 2015;27:55–60.
Liu Y, Wang H, Du N, et al. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob Agents Chemother. 2009;53:512–18.
Lulitanond A, Ito T, Li S, et al. ST9 MRSA strains carrying a variant of type IX SCCmec identified in the Thai community. BMC Infect Dis. 2013;13:214.
Wan MT, Lauderdale TS, Chou CC. Characteristics and virulence factors of livestock associated ST9 methicillin-resistant Staphylococcus aureus with a novel recombinant staphylocoagulase type. Vet Microbiol. 2013;162:779–84.
Wang D, Wang Z, Yan Z, et al. Bovine mastitis Staphylococcus aureus: antibiotic susceptibility profile, resistance genes and molecular typing of methicillin-resistant and methicillin-sensitive strains in China. Infect Genet Evol. 2015;31:9–16.
Lo YP, Wan MT, Chen MM, Su HY, Lauderdale TL, Chou CC. Molecular characterization and clonal genetic diversity of methicillin-resistant Staphylococcus aureus of pig origin in Taiwan. Comp Immunol Microbiol Infect Dis. 2012;35:513–21.
Moser A, Stephan R, Corti S, Johler S. Comparison of genomic and antimicrobial resistance features of latex agglutination test-positive and latex agglutination test-negative Staphylococcus aureus isolates causing bovine mastitis. J Dairy Sci. 2013;96:329–34.
Luzzago C, Locatelli C, Franco A, et al. Clonal diversity, virulence-associated genes and antimicrobial resistance profile of Staphylococcus aureus isolates from nasal cavities and soft tissue infections in wild ruminants in Italian Alps. Vet Microbiol. 2014;170:157–61.
Spoor LE, McAdam PR, Weinert LA, et al. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. MBio. 2013;4:e00356–13.
Lozano C, Gómez-Sanz E, Benito D, Aspiroz C, Zarazaga M, Torres C. Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int J Med Microbiol. 2011;301:500–5.
Egyir B, Guardabassi L, Esson J, et al. Insights into nasal carriage of Staphylococcus aureus in an urban and a rural community in Ghana. PLoS ONE. 2014;9:e96119.
Gómez-Sanz E, Torres C, Lozano C, et al. Detection, molecular characterization, and clonal diversity of methicillin-resistant Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Pathog Dis. 2010;7:1269–77.
Menegotto F, González-Cabrero S, Cubero Á, et al. Clonal nature and diversity of resistance, toxins and adhesins genes of meticillin-resistant Staphylococcus aureus collected in a Spanish hospital. Infect Genet Evol. 2012;12:1751–8.
Argudín MA, Argumosa V, Mendoza MC, Guerra B, Rodicio MR. Population structure and exotoxin gene content of methicillin-susceptible Staphylococcus aureus from Spanish healthy carriers. Microb Pathog. 2013;54:26–33.
Alba P, Feltrin F, Cordaro G, et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in European farmed animals: high genetic relatedness of isolates from Italian cattle herds and humans. PLoS ONE. 2015;10:e0137143.
Normanno G, Dambrosio A, Lorusso V, Samoilis G, Di Taranto P, Parisi A. Methicillin-resistant Staphylococcus aureus (MRSA) in slaughtered pigs and abattoir workers in Italy. Food Microbiol. 2015;51:51–6.
Agersø Y, Hasman H, Cavaco LM, Pedersen K, Aarestrup FM. Study of methicillin resistant Staphylococcus aureus (MRSA) in Danish pigs at slaughter and in imported retail meat reveals a novel MRSA type in slaughter pigs. Vet Microbiol. 2012;157:246–50.
Pilla R, Castiglioni V, Gelain ME, et al. Long-term study of MRSA ST1, t127 mastitis in a dairy cow. Vet Rec. 2012;170:312.
Buyukcangaz E, Velasco V, Sherwood JS, Stepan RM, Koslofsky RJ, Logue CM. Molecular typing of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) isolated from animals and retail meat in North Dakota, United States. Foodborne Pathog Dis. 2013;10:608–17.
Frana TS, Beahm AR, Hanson BM, et al. Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS ONE. 2013;8:e53738.
Molla B, Byrne M, Abley M, et al. Epidemiology and genotypic characteristics of methicillin-resistant Staphylococcus aureus strains of porcine origin. J Clin Microbiol. 2012;50:3687–93.
Khanna T, Friendship R, Dewey C, Weese JS. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol. 2008;128:298–303.
Lowder BV, Guinane CM, Ben Zakour NL, et al. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:19545–50.
García-Álvarez L, Holden MT, Lindsay H, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603.
Paterson GK, Harrison EM, Holmes MA. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014;22:42–7.
Becker K, Ballhausen B, Köck R, Kriegeskorte A. Methicillin resistance in Staphylococcus isolates: the “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int J Med Microbiol. 2014;304:794–804.
Porrero MC, Valverde A, Fernández-Llario P, et al. Staphylococcus aureus carrying mecC gene in animals and urban wastewater. Spain Emerg Infect Dis. 2014;20:899–901.
Kerschner H, Harrison EM, Hartl R, Holmes MA, Apfalter P. First report of mecC MRSA in human samples from Austria: molecular characteristics and clinical data. New Microbes New Infect. 2014;3:4–9.
Espinosa-Gongora C, Harrison EM, Moodley A, Guardabassi L, Holmes MA. MRSA carrying mecC in captive mara. J Antimicrob Chemother. 2015;70:1622–4.
Gómez P, Lozano C, Camacho MC, Lima-Barbero JF, Hernández JM, Zarazaga M, Höfle Ú, Torres C. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J Antimicrob Chemother. 2015.
Gómez P, Lozano C, González-Barrio D, Zarazaga M, Ruiz-Fons F, Torres C. High prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in a semi-extensive red deer (Cervus elaphus hispanicus) farm in Southern Spain. Vet Microbiol. 2015;177:326–31.
Porrero MC, Mentaberre G, Sánchez S, et al. Carriage of Staphylococcus aureus by free-living wild animals in Spain. Appl Environ Microbiol. 2014;80:4865–70.
Haenni M, Châtre P, Dupieux C, et al. mecC-positive MRSA in horses. J Antimicrob Chemother. 2015;70:3401–2.
Harrison EM, Paterson GK, Holden MT, et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med. 2013;5:509–15.
Haenni M, Châtre P, Tasse J, et al. Geographical clustering of mecC-positive Staphylococcus aureus from bovine mastitis in France. J Antimicrob Chemother. 2014;69:2292–3.
Harrison EM, Paterson GK, Holden MT, et al. A Staphylococcus xylosus isolate with a new mecC allotype. Antimicrob Agents Chemother. 2013;57:1524–8.
Harrison EM, Paterson GK, Holden MT, et al. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J Antimicrob Chemother. 2014;69:911–8.
Loncaric I, Kübber-Heiss A, Posautz A, et al. J. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. Antimicrob Chemother. 2013;68:2222–5.
Armand-Lefevre L, Ruimy R, Andremont A. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg Infect Dis. 2005;11:711–4.
Devriese LA, Van Damme LR, Fameree L. Methicillin (cloxacillin)-resistant Staphylococcus aureus strains isolated from bovine mastitis cases. Zentralbl. Veterinarmed. 1972; B 19, 598–605
Ferber D. Infectious disease. From pigs to people: the emergence of a new superbug. Science. 2010;329(5995):1010–1.
Denis O, Suetens C, Hallin M, et al. Methicillin-resistant Staphylococcus aureus ST398 in swine farm personnel. Belgium Emerg Infect Ds. 2009;15:1098–101.
Bens CC, Voss A, Claassem CH. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J Clin Microbiol. 2006;44:1875–6.
Argudín MA, Rodicio MR, Guerra B. The emerging methicillin-resistant Staphylococcus aureus ST398 clone can easily be typed using the Cfr9I SmaI-neoschizomer. Lett Appl Microbiol. 2010;50:127–30.
Rasschaert G, Vanderhaeghen W, Dewaele I, et al. Comparison of fingerprinting methods for typing methicillin-resistant Staphylococcus aureus sequence type 398. J Clin Microbiol. 2009;47:3313–22.
Aarestrup FM, Cavaco L, Hasman H. Decreased susceptibility to zinc chloride is associated with methicillin resistant Staphylococcus aureus CC398 in Danish swine. Vet Microbiol. 2010;142:455–7.
Cavaco LM, Hasman H, Aarestrup FM. Zinc resistance of Staphylococcus aureus of animal origin is strongly associated with methicillin resistance. Vet Microbiol. 2011;150:344–8.
Cavaco LM, Hasman H, Stegger M, et al. Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrob Agents Chemother. 2010;54:3605–8.
Li S, Skov RL, Han X, et al. Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2011;55:3046–50.
Slifierz MJ, Friendship R, Weese JS. Zinc oxide therapy increases prevalence and persistence of methicillin-resistant Staphylococcus aureus in pigs: a randomized controlled trial. Zoonoses Public Health. 2015;62:301–8.
Slifierz MJ, Friendship RM, Weese JS. Methicillin-resistant Staphylococcus aureus in commercial swine herds is associated with disinfectant and zinc usage. Appl Environ Microbiol. 2015;81:2690–5.
Moodley A, Nielsen SS, Guardabassi L. Effects of tetracycline and zinc on selection of methicillin-resistant Staphylococcus aureus (MRSA) sequence type 398 in pigs. Vet Microbiol. 2011;152:420–3.
Smith TC, Male MJ, Harper AL, et al. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One. 2009;4:e4258.
Leedom Larson KR, Harper AL, Hanson BM, et al. Methicillin-resistant Staphylococcus aureus in pork production shower facilities. Appl Environ Microbiol. 2011;77:696–8.
Smith TC, Gebreyes WA, Abley MJ, et al. Methicillin-resistant Staphylococcus aureus in pigs and farm workers on conventional and antibiotic-free swine farms in the USA. PLoS ONE. 2013;8:e63704.
Nadimpalli M, Rinsky JL, Wing S, et al. Persistence of livestock-associated antibiotic-resistant Staphylococcus aureus among industrial hog operation workers in North Carolina over 14 days. Occup Environ Med. 2015;72:90–9.
Rinsky JL, Nadimpalli M, Wing S, et al. Livestock-associated methicillin and multidrug resistant Staphylococcus aureus is present among industrial, not antibiotic-free livestock operation workers in North Carolina. PLoS ONE. 2013;8:e67641.
Osadebe LU, Hanson B, Smith TC, Heimer R. Prevalence and characteristics of Staphylococcus aureus in Connecticut swine and swine farmers. Zoonoses Public Health. 2013;60:234–43.
Dressler AE, Scheibel RP, Wardyn S, et al. Prevalence, antibiotic resistance and molecular characterisation of Staphylococcus aureus in pigs at agricultural fairs in the USA. Vet Rec. 2012;170:495.
Hawken P, Weese JS, Friendship R, Warriner K. Longitudinal study of Clostridium difficile and Methicillin-resistant Staphylococcus aureus associated with pigs from weaning through to the end of processing. J Food Prot. 2013;76:624–30.
Weese JS, Zwambag A, Rosendal T, Reid-Smith R, Friendship R. Longitudinal investigation of methicillin-resistant Staphylococcus aureus in piglets. Zoonoses Public Health. 2011;58:238–43.
Gordoncillo MJ, Abdujamilova N, Perri M, Donabedian S, Zervos M, Bartlett P. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in backyard pigs and their owners, Michigan, USA. Zoonoses Public Health. 2012;59:212–6.
Neela V, Mohd ZA, Mariana NS, van Belkum A, Liew YK, Rad EG. Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J Clin Microbiol. 2009;47:4138–40.
Anukool U, O’Neill CE, Butr-Indr B, Hawkey PM, Gaze WH, Wellington EM. Meticillin-resistant Staphylococcus aureus in pigs from Thailand. Int J Antimi-Crob Agents. 2011;38:86–7.
Tsai HY, Liao CH, Cheng A, et al. Isolation ofmeticillin-resistant Staphylococcus aureus sequence type 9 in pigs in Taiwan. Int J Antimicrob Agents. 2012;39:449–51.
Fang HW, Chiang PH, Huang YC. Livestock-associated methicillin-resistant Staphylococcus aureus in pigs and related personnel in Taiwan. PLoS ONE. 2014;9:e88826.
Cui S, Li J, Hu C, et al. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J Antimicrob Chemother. 2009;64:680–3.
Yan X, Yu X, Tao X, et al. Staphylococcus aureus ST398 from slaughter pigs in northeast China. Int J Med Microbiol. 2014;304:379–83.
Ho PL, Siu JT, Law PY, et al. Clonality and antimicrobial susceptibility of Staphylococcus aureus and methicillin-resistant S. aureus isolates from food animals and other animals. J Clin Microbiol. 2012;50:3735–7.
Vestergaard M, Cavaco LM, Sirichote P, et al. SCCmec type IX element in methicillin-resistant Staphylococcus aureus spa type t337 (CC9) isolated from pigs and pork in Thailand. Front Microbiol. 2012;3:103.
Wagenaar JA, Yue H, Pritchard J, et al. Unexpected sequence types in livestock associated methicillin- resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet Microbiol. 2009;139:405–9.
Boost MV, Wong A, Ho J, O’Donoghue M. Isolation of methicillin-resistant Staphylococcus aureus (MRSA) from retail meats in Hong Kong. FoodbornePathog Dis. 2013;10:705–10.
Guardabassi L, O’Donoghue M, Moodley A, Ho J, Boost M. Novel lineage of methicillin-resistant Staphylococcus aureus, Hong Kong. Emerg Infect Dis. 2009;15:1998–2000.
Larsen J, Imanishi M, Hinjoy S, et al. Methicillin-resistant Staphylococcus aureus ST9 in pigs in Thailand. PLoS ONE. 2012;7:e31245.
Sergio DM, Koh TH, Hsu LY, Ogden BE, Goh AL, Chow PK. Investigation of methicillin-resistant Staphylococcus aureus in pigs used for research. J MedMicrobiol. 2007;56:1107–9.
Baba K, Ishihara K, Ozawa M, Tamura Y, Asai T. Isolation of meticillin-resistant Staphylococcus aureus (MRSA) from swine in Japan. Int J Antimicrob Agents. 2010;36:352–4.
El-Jakee JK, Atta N, Samy A, Bakry MEAE, Kandil M, GadEl-Said W. Antimicrobial resistance in clinical isolates of Staphylococcus aureus from bovine and human sources in Egypt. Glob Vet. 2011;7:581–6.
El Seedy FR, El Hakim AS, Syame SF, Osman NM. Advanced techniques used for isolation and characterization of Staphylococcus aureus isolated from mastitic buffaloes. Glob Vet. 2012;8:144–52.
Fall C, Seck A, Richard V, et al. Epidemiology of Staphylococcus aureus in pigs and farmers in the largest farm in Dakar, Senegal. Foodborne Pathog Dis. 2012;9:962–5.
Gharsa H, BenSlama K, Lozano C, et al. Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Vet Microbiol. 2012;156:367–73.
Ben Slama K, Gharsa H, Klibi N, et al. Nasal carriage of Staphylococcus aureus in healthy humans with different levels of contact with animals in Tunisia: genetic lineages, methicillin resistance, and virulence factors. Eur J Clin Microbiol Infect Dis. 2011;30:499–508.
Roberts GA, Houston PJ, White JH, et al. Impact of target site distribution for Type I restriction enzymes on the evolution of methicillin-resistant Staphylococcus aureus (MRSA) populations. Nucleic Acids Res. 2013;41:7472–84.
Lindsay JA. Genomic variation and evolution of Staphylococcus aureus. Int J Med Microbiol. 2010;300:98–103.
Tulinski P, Fluit AC, Wagenaar JA, Mevius D, van de Vijver L, Duim B. Methicillin-resistant coagulase-negative staphylococci on pig farms as a reservoir of heterogeneous staphylococcal cassette chromosome mec elements. Appl Environ Microbiol. 2012;78:299–304.
Kern A, Perreten V. Clinical and molecular features of methicillin-resistant, coagulase-negative staphylococci of pets and horses. J Antimicrob Chemother. 2013;68:1256–66.
Park YK, Paik YH, Yoon JW, Fox LK, Hwang SY, Park YH. Dissimilarity of ccrAB gene sequences between methicillin-resistant Staphylococcus epidermidis and methicillin-resistant Staphylococcus aureus among bovine isolates in Korea. J Vet Sci. 2013;14:299–305.
Grøntvedt C, Sunde M, Angen Ø, Steihaug Barstad A, Åmdal S, Løtvedt S, Elstrøm P, Wik Larssen K, Stegger M, Skov R, Fredriksen B, Karlsen O, Hofmo P, Urdahl A. Control of LA-MRSA in swine - is it possible? Lessons learned from outbreaks and eradication in Norway. 4th ASM-ESCMID Conference on methicillin-resistant staphylococci in animals: veterinary and public health implications November 2–5, 2015, Chicago Illinois. Page 27 S4:2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Dr. Butaye, Dr. Argudín, and Dr. Smith have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Bacteriology
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Butaye, P., Argudín, M.A. & Smith, T.C. Livestock-Associated MRSA and Its Current Evolution. Curr Clin Micro Rpt 3, 19–31 (2016). https://doi.org/10.1007/s40588-016-0031-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40588-016-0031-9