Abstract
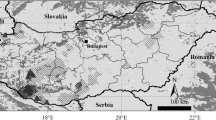
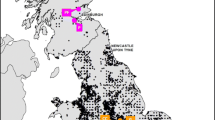
Common hamsters Cricetus cricetus (L.)show a highly fragmented distributionpattern across Europe. Over the last decades,human influence caused significant populationdeclines in particular at the western rangeboundary. Despite the initiation of breedingand release programs the genetic structure anddiversity of European common hamsterpopulations is largely unknown. In this study,hamsters from ten localities in five Europeancountries were investigated. Mitochondrialcontrol region was sequenced from 145 animalsrepresenting all sampled populations. 385hamster were screened for polymorphisms at 11microsatellite loci. Both marker systemsrevealed extensive genetic differentiationamong European common hamsters. Westernpopulations displayed very low levels of mtDNAdiversity (H = 0 − 0.2, Alsace, Limburg,Flanders, Baden-Wuerttemberg) compared toeastern populations from Saxony-Anhalt,Thuringia and Southern Moravia (H = 0.663− 0.816). Microsatellite analyses revealed asimilar pattern with low to moderate diversityvalues in western hamsters (A = 1.636 −5.364; H e = 0.111 − 0.504) and highlevels of polymorphism in eastern hamsters(A = 8.909 − 9.818; H e = 0.712− 0.786). High microsatellite based F STmeasures (up to 0.635) suggest a typical islandmodel of distribution with no current gene flowbetween most areas. Western hamster populationsexhibit obvious similarities in mitochondrialhaplotype and microsatellite alleledistributions. Gene trees group westernhamsters consistently together on the samebranch but bootstrap values never reachedsignificance. There are strong indications thatlow diversity in western populations ispartially caused by a joint historic founderevent and not only by recent population breakdowns. Overlapping mitochondrial haplotypesprove a close association between westernhamsters and animals from the east German rangein the recent past which does not support theexistence of a separate subspecies C. c.canescens in Europe. Hamsters from southernMoravia emerged as the genetically mostdistinguished population and could be part of a different genetic lineage in Europe.
Similar content being viewed by others
References
Banks M, Eichert W (1999) WhichRun. A Computer Program for Population Assignment of Individuals Based on Multilocus Genotype Data, Version 4.0. Bodega Marine Laboratory, University of California, Davis.
Barrett EM, Gurnell J, Malarky G et al. (1999) Genetic structure of fragmented populations of red squirrel (Sciurus vulgaris) in the UK. Molecular Ecology, 8, S55–S63.
Berdyugin KI, Bolshakov VN (1998) The common hamster (Cricetus cricetus L.) in the eastern part of the area. In: Ecology and Protection of the Common Hamster (eds. Stubbe M, Stubbe A), pp. 43–80. Wissenschaftliche Beiträge Martin-Luther-Universität Halle-Wittenberg, Germany.
Calinescu RJ (1931) Ñber Verbreitung und Einfälle von Cricetus cricetus nehringi MTSCH. in Rumänien. Zeitschrift für Säugetierkunde, 6, 231–233.
Charlesworth B (1998) Measures of divergence between populations and the effect of forces that reduce variability. Molecular Biology and Evolution, 15, 538–543.
Clement M, Posada D, Crandall KA (2000) TCS: A computer program to estimate gene genealogies. Molecular Ecology, 9, 1657–1659.
Cornuet JM, Luitkart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics, 144, 2001–2014.
Effenberger S, Suchentrunk F (1999) RFLP analysis of the mitochondrial DNA of otters (Lutra lutra) from Europe-implications for conservation of a flagship species. Biological Conservation, 90, 229–234.
Ehrich D, Fedorov VB, Stenseth NC et al. (2000) Phylogeography and mitochondrial DNA (mtDNA) diversity in North American collared lemmings (Dicrostonyx groenlandicus). Molecular Ecology, 9, 329–337.
Ehrich D, Jorde PE, Krebs CJ et al. (2001) Spatial structure of lemming populations (Dicrostonyx groenlandicus) fluctuating in density. Molecular Ecology, 10, 481–495.
Ellegren H, Hartmann G, Johansson M, Andersson L (1993) Major histocompatibility complex monomorphism and low levels of DNA fingerprinting variability in a reintroduced and rapidly expanding population of beavers. Proc. Natl. Acad. Sci. USA, 90, 8150–8153.
Estoup A, Tailliez C, Cornuet J-M et al. (1995) Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae). Molecular Biology and Evolution, 12, 1074–1084.
Estoup A, Jarne P, Cornuet J-M (2002) Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Molecular Ecology 11, 1591–1604.
Faulkes CG, Abbott DH, O'Brian et al. (1997) Micro-and macrogeographical genetic structure of naked mole-rats Heterocephalus glaber. Molecular Ecology, 6, 615–628.
Gerber AS, Templeton AR (1996) Population sizes and within-deme movement of Trimerotropis saxatilis (Acrididae), a grasshopper with a fragmented distribution. Oecologia, 105, 343–350.
Gerlach G, Musolf K (2000) Fragmentation of landscape as a cause for genetic subdivision in bank voles. Conservation Biology, 14, 1066–1074.
Goudet J (1995) FSTAT (vers. 2.1): A computer program to calculate F-statistics. J. Heredity, 86, 485–486.
Grulich I (1987) Variability of Cricetus cricetus in Europe. Acta Sc. Nat. Brno, 21, 1–53.
Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportions for multiple alleles. Biometrics, 48, 361–372.
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society, 58, 247–276.
Karaseva EV (1962) A study of the pecularities of territory utilization by the hamster in the Altai territory carried out with the use of labeling. Zoological Journal (Russia), 41, 275–284.
Koh HS, Lee W-J, Kocher TD (2000) The genetic relationship of two subspecies of striped field mice, Apodemus agrarius coreae and Apodemus agrarius chejuensis. Heredity, 85, 30–36.
Kumar S, Tamura K, Jakobsen IB et al. (2001) MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics, 17, 1244–1245.
Laikre L, Ryman N (1991) Inbreeding depression in a captive wolf (Canis lupus) population. Conservation Biology, 5, 33–40.
Leijs R, Apeldoorn RC, Bijlsma R (1999) Low genetic differentiation in north-west European populations of the locally endangered root vole, Microtus oeconomus. Biological Conservation, 87, 39–48.
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Research, 27, 209–220.
Mitchell-Jones AJ, Amori G, Bogdanowicz W et al. (1999) The Atlas of European Mammals. Poyser, London.
Moritz C (1994) Defining 'evolutionarily significant units' for conservation. Trends in Ecology and Evolution, 9, 373–375.
Nakamichi N, Rhoads DD, Hayashi JI, Kagawa Y, Matsumura T (1998) Detection, localization, and sequence analyses of mitochondrial regulatory region RNAs in several mammalian species. J. Biochem., 123, 392–398.
Nei M (1972) Genetic distances between populations. American Naturalists, 106, 283–292.
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA, 70, 3321–3323.
Neumann K, Maak S, Stuermer IW et al. (2001) Low microsatellite variation in laboratory gerbils. J. Heredity, 92, 71–74.
Niethammer J (1982) Cricetus cricetus (Linnaeus, 1758)-Hamster (Feldhamster). In: Handbuch der Säugetiere Europas (eds. Niethammer J, Krapp F), pp. 7–28. Akad. Verlagsgesellschaft-Wiesbaden.
Oli MK, Holler NR, Wooten MC (2001) Viability analysis of endangered Gulf Coast beach mice (Peromyscus polionotus) populations. Biological Conservation, 97, 107–118.
Page RDM (1996) TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences, 12, 357–358.
Petzsch H (1950) Der Hamster. Neue Brehm-Bücherei. Wittenberg-Lutherstadt.
Raymond M, Rousset F (1995) GENEPOP (Version 1.2). Population genetics software for exact tests and ecumenicism. J. Heredity, 86, 248–249.
Rozas J, Rozas R (1999) DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics, 15, 174–175.
Schneider S, Roessli D, Excoffier L (2000) Arlequin, Version 2.0: A Software for Population Genetic Data Analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva.
Spitzenberger F, Bauer K (2001) Hamster Cricetus cricetus (Linnaeus, 1758). In: Die Säugetierfauna Österreichs (ed. Spitzenberger F), pp. 406–415. Bundesministerium für Landund Forstwirtschaft, Umwelt und Wasserwirtschaft, Band 13.
Tallmon DA, Draheim HM, Scott-Mills L, Allendorf FW (2002) Insights into recently fragmented vole populations from combined genetic and demographic data. Molecular Ecology, 11, 699–709.
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. Genetics, 132, 619–633.
Vohralik V (1974) Biology of the reproduction of the common hamster Cricetus cricetus (L.). Vestnik Ceskoslovenske Spolecnosti Zoologicke, 38, 228–240.
Vohralik V, Andera M (1976) Distribution of the common hamster, Cricetus cricetus (L.) in Czechoslovakia. Lynx, 18, 85–97.
Walker CW, Vilà C, Landa A et al. (2001) Genetic variation and population structure in Scandinavian wolverine (Gulo gulo) populations. Molecular Ecology, 10, 53–63.
Weinhold U (1996) Radiotelemetrische Untersuchungen zum Raum-Zeitverhalten des Feldhamsters (Cricetus cricetus L., 1758) auf landwirtschaftlich genutzten Flächen im Raum Mannheim-Heidelberg. Säugetierkundliche Informationen, 20, 129–134.
Zenger KR, Richardson BJ, Vachot-Griffi AM (2003) A rapid population expansion retains genetic diversity within European rabbits in Australia. Molecular Ecology, 12, 789–794.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Neumann, K., Jansman, H., Kayser, A. et al. Multiple bottlenecks in threatened western European populations of the common hamster Cricetus cricetus (L.). Conservation Genetics 5, 181–193 (2004). https://doi.org/10.1023/B:COGE.0000030002.01948.b3
Issue Date:
DOI: https://doi.org/10.1023/B:COGE.0000030002.01948.b3