Abstract
Purpose
F13640 (a.k.a. befiradol, NLX-112) is a highly selective 5-HT1A receptor ligand that was selected as a PET radiopharmaceutical-candidate based on animal studies. Due to its high efficacy agonist properties, [18F]F13640 binds preferentially to functional 5-HT1A receptors, which are coupled to intracellular G-proteins. Here, we characterize brain labeling of 5-HT1A receptors by [18F]F13640 in humans and describe a simplified model for its quantification.
Methods
PET/CT and PET-MRI scans were conducted in a total of 13 healthy male volunteers (29 ± 9 years old), with arterial input functions (AIF) (n = 9) and test–retest protocol (n = 8). Several kinetic models were compared (one tissue compartment model, two-tissue compartment model, and Logan); two models with reference region were also evaluated: simplified reference tissue model (SRTM) and the logan reference model (LREF).
Results
[18F]F13640 showed high uptake values in raphe nuclei and cortical regions. SRTM and LREF models showed a very high correlation with kinetic models using AIF. As concerns test–retest parameters and the prolonged binding kinetics of [18F]F13640, better reproducibility, and reliability were found with the LREF method. Cerebellum white matter and frontal lobe white matter stand out as suitable reference regions.
Conclusion
The favorable brain labeling and kinetic profile of [18F]F13640, its high receptor specificity and its high efficacy agonist properties open new perspectives for studying functionally active 5-HT1A receptors, unlike previous radiopharmaceuticals that act as antagonists. [18F]F13640’s kinetic properties allow injection outside of the PET scanner with delayed acquisitions, facilitating the design of innovative longitudinal protocols in neurology and psychiatry.
Trial Registration.
Trial Registration EudraCT 2017–002,722-21.
Similar content being viewed by others
Introduction
Serotonin (5-hydroxytryptamine, 5-HT) is known to have a variety of functions in the central nervous system, which are mediated by a diversity of receptors. Among them, the serotonin 1A receptor subtype (5-HT1A) is a G-coupled protein receptor (GCPR) which has attracted extensive interest because it is involved in regulation of mood, cognition, pain, and movement [1]. Consequently, various positron emission tomography (PET) radiotracers have been developed to target 5-HT1A receptors, including [O-methyl-11C]WAY100635 [2], [carbonyl-11C]WAY100635 [2, 3], and [18F]MPPF, a fluorinated derivate of WAY100635 [4, 5]. [18F]MPPF binding is observed mainly in brain regions with a high density of 5-HT1A receptors such as hippocampus and raphe nuclei [5,6,7] and it has been used to explore various psychiatric and neurologic diseases such as epilepsy [8], narcolepsy [9], Alzheimer’s disease [10], or multiple system atrophy [11]. Other derivatives of WAY100635 have been also proposed as radiopharmaceuticals but have been less used in human [12].
Nevertheless, although [11C]WAY100635 and [18F]MPPF are widely used for in vivo exploration of 5-HT1A receptors, PET imaging using these radiopharmaceuticals remains limited in terms of pathophysiological interpretation because of their antagonist pharmacological properties. Indeed, antagonists bind both G protein-coupled and G-protein uncoupled receptors with the same affinity, labeling the total receptor population, regardless of its functional status. In contrast, agonists have higher affinity for their target GPCRs when the latter are coupled to G-proteins, i.e., they are in a functional state which is directly associated with neurotransmission [13, 14]. Thus, we hypothesized that highly specific 5-HT1A receptor agonist radiotracers would constitute useful tools to explore endogenous serotonergic neurotransmission and pathophysiological changes that specifically affect functional receptors and which would not be detectable using antagonist radiopharmaceuticals [15].
Very few 5-HT1A agonists have been used as chemical templates to develop a PET radiopharmaceutical. [11C]CUMI-101, which was initially presented as an agonist, was later found to act as a partial agonist or even as an antagonist [16], and it also binds to α1 adrenoceptors [17]. These suboptimal pharmacological properties explain its modest sensitivity to pharmacological challenges or to endogenous serotoninergic changes in human [18, 19]. In this context, we chose to use highly specific and pharmacologically well-characterized 5-HT1A receptor agonists to develop the corresponding radiopharmaceuticals. Following several studies, we selected F13640 (a.k.a. befiradol or NLX-112), which possesses high affinity (nanomolar Ki) for 5-HT1A receptors, high selectivity (> 1000-fold) over a large range of other CNS targets and whose chemical structure includes a fluorine which is substitutable by a fluorine-18. As a result, [18F]F13640 was proposed as the first preclinical fluorinated 5-HT1A receptor agonist radiopharmaceutical, supported by compelling preclinical data in animal models [20, 21]. Although a first image obtained in a healthy volunteer suggested favorable brain penetration by [18F]F13640 [22], it remained to be demonstrated whether [18F]F13640 could become a usable radiopharmaceutical for future clinical investigation. The objectives of the present study were, therefore, to perform a full PET kinetic modeling of [18F]F13640 using arterial input function (AIF), to identify a reference region suitable for a simplified modeling method, and to assess reproducibility with test–retest scans. Ultimately, the present data characterizing [18F]F13640 as a selective agonist radiopharmaceutical will, for the first time, enable the investigation of changes in both 5-HT1A receptor expression and functionality in patients suffering from disorders arising from dysfunctional serotonergic neurotransmission.
Material and methods
Synthesis and quality control
[18F]F13640 (3-chloro-4-fluorophenyl)-[4-18fluoro-4-[[(5-methylpyridin-2-yl)methylamino]methyl]piperidin-1-yl]methanone; a.k.a. [18F]NLX-112 or [18F]befiradol) was synthesized as previously described [21]. Briefly, radiolabeling was obtained by a nucleophilic fluoro-substitution on the F13640 nitro precursor using an automated radiosynthesizer (Neptis, ORA). Chemical and radiochemical purity measured by HPLC was higher than 95%. Mean molar activity (EOS) was 82 ± 18 GBq/µmol.
Study design
The study was performed according to the ethical standards of the institutional and national research committee and with the principles of the 1964 Declaration of Helsinki. The study was approved by a French ethics committee (Eudra-CT: 2017–002,722-21) and pre-registered on the ClinicalTrials.gov database (NCT03347331).
Participants were healthy volunteers, i.e., without neurological or psychiatric disorders, active infectious disease, severe and progressive medical pathology, without addiction (smoking, cannabis, alcohol…), or MRI and PET contraindications. Twenty volunteers were screened in the study and signed consent. Two volunteers failed inclusion due to exclusion criteria (MRI contraindication and body weight superior to 90 kg). The first eight volunteers were included in a pilot study in which the subjects underwent an anatomical MRI scan (3D T1-weighted sequence on a 1.5-T Siemens Magneton scanner, Siemens AG, Erlanger, Germany) and a 90 min PET/CT (Siemens Biograph mCT64) scan with arterial input function (AIF, data not shown). The later 10 volunteers were included to perform a test–retest protocol with two PET-MRI scans as described below. Four participants had AIF measurements on one of the two visits. Two volunteers were lost to follow-up. To sum up, data analysis focused on four participants when concerning PET modeling with AIF and eight participants for test–retest analysis. Figure 1 summarizes recruitment of healthy subjects in the study.
PET-MRI test–retest protocol
Participants underwent a PET-MRI acquisition on the Siemens mMR Biograph system. Because the pilot study group scans showed that [18F]F13640 activity curves slowly continued to vary even at later time points, suggesting that there may be further changes beyond 90 min. A PET-MRI acquisition of almost 4 h was tested to observe kinetics for a longer period. For the comfort of the subjects, the acquisition was carried out in two parts. In part 1 (PET1), list-mode PET data were acquired for 90 min directly after the injection of [18F]F13640 (150 MBq + 1 MBq/kg ± 10%) (PET1, [0; 90] min. post injection). Subjects were then taken out of the camera for a break. One hour later, for part 2 (PET2), subjects had a second PET-MRI scan for 75 min (PET2; [150; 225] min. post injection). Participants performed this PET-MRI protocol twice (test and retest sessions) 1 to 9 weeks apart.
During PET1, a 3D T1 MPRAGE MRI was acquired in sagittal orientation, with a matrix size of 256 × 256 × 176 and a voxel size of 1 mm iso. TR/TE was 3300/2.45 ms, TI 1100 ms, and flip angle 8°. A quicker T1 MPRAGE MRI was acquired at the beginning of PET2 to accurately register data from the two sessions (sagittal acquisition, matrix size 256 × 256 × 176, voxel size 1 mm iso, TR/TE 1800/2.34 ms, TI = 850 ms, flip angle 8).
PET-MRI image processing
The PET data from the two parts were reconstructed independently. First, a MR-based attenuation correction map was generated from the T1 MPRAGE of each part [23]. PET1 and PET2 list modes were then corrected for motion with the EBER algorithm [24]. PET1 was rebinned into 24 frames of variable duration (8 × 15 s, 3 × 60 s, 5 × 120 s, 1 × 300 s, 7 × 600 s), and PET2 was rebinned into 8 frames (7 × 600 s, 1 × 300 s). Sinograms were normalized and corrected for attenuation [23], scatters, and randoms. PET reconstructions were performed with Siemens e7tools using the OP-OSEM algorithm with PSF, 3 iterations, and 21 subsets. A matrix size of 256 and a zoom of 3 were applied yielding a voxel size of 0.93 × 0.93 × 2.03 mm with a 4 mm 3D post-reconstruction gaussian filtering.
Both dynamic PET series from PET1 and PET2 were combined in a single dynamic series in the following way. The mean of each dynamic PET series was computed. The mean of PET2 was rigidly coregistered to the mean of PET1. This rigid transformation was applied to all frames of PET2 to obtain the PET2 frames aligned with the PET1 frames. Decay correction was applied to PET2 and PET1 setting the reference time for both series to the start time of the PET1 scan. PET1 and PET2 were finally concatenated in a single and harmonized dynamic 4D file. These preprocessing steps were performed with the minc toolkit functions (http://bic-mni.github.io).
In addition to the above, the T1 image of the retest session and the T1 image of the test session were coregistered by applying the computed rigid coregistration matrix to all images of the retest session. The structural T1 image from the test session was automatically segmented into anatomical regions using the multi-atlas propagation with enhanced registration (MAPER) method [25] and the 95-region Hammersmith atlas [26,27,28]. The raphe nucleus was segmented based on functional data from a previous study [29]. The regional segmentation was projected to test and retest sessions, and regional time activity curves (TAC) were extracted for a selection of brain regions. Regions selected for analysis were cortical regions (cingulate, frontal, occipital, parietal, temporal superior, and temporal inferior), amygdala, central grey nuclei, hippocampus, insula, parahippocampal gyrus, thalamus, brainstem, dorsal and median raphe nuclei, cerebellum (total, grey matter, and white matter), vermis, frontal lobe white matter, and corpus callosum.
Arterial input function (AIF)
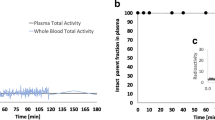
Four participants underwent AIF measures during their test or retest session. AIF, free fraction, and metabolites were measured based on 29 blood samples. Samples were collected manually after arterial catheterization with local anesthesia (lidocaine patch 5%). Sample time points were as follows: every 5 s in the first minute, every 15 s in the second minute, every 30 s in the third minute, and at time 5, 10, 20, 30, 40, 60, 75, 90, 160, and 205 min. Whole blood radioactivity was counted on every sample using a gamma counter (Perkin-Elmer) to measure the whole blood curve (Cwb). Plasma was collected and counted after centrifugation (4 min, 3000 G at 4 °C) on 18 out of 29 samples (t = 15 s and every sample after the first minute to calculate plasma to whole blood ratio fwb). Uncorrected plasma curve (Cp) was determined by the mean fwb and the whole blood curve: Cp (t) = fwb.Cwb (t). On 5 samples (t = 2, 10, 30, 90, and 205 min) free fraction and metabolites were determined. For metabolites, 500 µL of plasma was added to 750 µL of acetonitrile with cold carrier of F13640 at 20 mg/L, centrifuged (4 min, 3000 G at 4 °C), filtered at 0.45 µm, diluted with water, and injected in a C8 HPLC column with a mixed mobile phase water/acetonitrile/TFA (60/40/0.1). Metabolites and [18F]F13640 were separated, and fractions were collected and counted in the gamma counter. The activity of the [18F]F13640 fraction was divided by the total activity recovered from the gamma counter to give the plasma parent fraction of unmetabolized [18F]F13640 (PPf). For plasma free fraction (fp), 1 mL of plasma was centrifugated (Centrifree®, Millipore) for 20 min, 2000 g at 20–25 °C, and 100 µL of ultrafiltrate plasma was counted in a gamma counter. After counting, all samples were weighed, and counts were corrected. The fp was calculated from the ratio of concentrations in the ultrafiltrate and whole plasma. The AIF was derived from Cp (t) according to: AIF (t) = PPf (t).Cp (t).fp (t).
Kinetic modeling
Cerebral TACs (PET (t)) were modeled with three different AIF models for participants who performed AIF (n = 4): one-tissue compartment (1TC), two-tissue compartment (2TC), and the Logan graphical method (LOGAN) [30]. Blood volume in tissue (Vb) was included as a model parameter in the operational equation according to: PET (t) = Vb.Cwb (t) x + (1-Vb).CT (t), where CT is the TAC in tissue. Model fit accuracies were compared using the Akaike Information Criterion (AIC). Two models with reference region were also assessed: the Simplified Reference Tissue Model (SRTM) [31] and the logan reference model (LREF) [30]. Four different reference regions were compared: corpus callosum (CC), cerebellum (CER), white matter cerebellum (CERWM), and frontal lobe white matter (FLWM). Distribution volume ratio (DVR) for models with AIF (DVR1TC or DVR2TC) was calculated as the ratio between the Vt of the region of interest and the Vt of the reference region. DVRs for SRTM (DVRSRTM) were calculated by adding one to the binding potential (BP) value. DVRs calculated with AIF models were compared to DVRs obtained with reference region models using linear regression to determine fitting parameters: intercept, slope, and determination coefficient (R2). Kinetic modeling was done using the Turku PET center utilities library (TPCCLIB, https://gitlab.utu.fi/vesoik/tpcclib).
Test–retest reproducibility and reliability
For all the participants in the test–retest study (n = 8), models with reference region were applied on both sessions. Bias and variability (VAR) were calculated to assess reproducibility, and the intraclass correlation coefficient (ICC) was computed to estimate reliability. Bias was calculated as (DVRretest − DVRtest)/DVRtest × 100 and VAR as the standard deviation (SD) of the bias. Parameters were expressed as percentage. ICC was calculated as (BSMSS-WSMSS)/(BSMSS + WSMSS) where the BSMSS is the mean sum of the square between subjects and WSMSS is the mean sum of the square within subjects [32].
Statistical analysis
Statistical analysis was performed using RStudio (RStudio Team 2020, http://www.rstudio.com/). Paired Student’s t-tests were used to assess differences between injected doses and molar activities between test and retest sessions. Significant threshold was set at p < 0.05.
Results
Subject demographics
Mean age of the participants was 29 ± 9 years in the test–retest study. No significant differences were found in the activity of the [18F]F13640 doses administered to the subjects or in the molar activities of [18F]F13640 between the test and retest (p value > 0.05). Due to an injection error, participant 8 received a dose of [18F]F13640 for a retest session that was slightly lower than the recommended dosages described in the study. Details are presented in Table 1.
Modeling study
Mean plasma parent fractions were 99.40% ± 0.00%, 97.40% ± 0.01%, 96.20% ± 0.01%, 95.40% ± 0.01%, and 95.30% ± 0.01% at 2, 10, 30, 90, and 205 min respectively after injection. One value was discarded for participant 2 at 90 min due to non-interpretable value. Mean plasma parent fraction was modeled with a one-exponential function:
with A0 = 0.046 and T = 9.06 min
Free plasmatic fraction was not constant over time and mean values were 0.48% ± 0.09%, 0.82% ± 0.08%, 1.14% ± 0.26%, 1.90% ± 0.71% and 1.61% ± 0.29% at 2, 10, 30, 90, and 205 min. Ratio plasma to whole blood was stable over time and mean value was 1.79 ± 0.03. Table 2 summarizes the pharmacokinetic parameters for each subject. Figure 2 shows an example of AIF corrected for metabolites, whole blood, and uncorrected plasma curves. Individual TACs and AIF are presented in online resource 1 (Fig S1, S2, S3, and S4).
Mean AIC using the one-tissue compartment model (1TC) was 320.88 ± 13.66 and 291.92 ± 25.46 for the two-tissue compartment model (2TC). Two-tissue compartment model better fits with the pharmacokinetic of [18F]F13640 when comparing the AIC but results in nonsensical numerical kinetics parameters (k3 and k4). Thus, the 1TC model was considered as the reference model for the results presented below. Fitted curves for 1TC and 2TC modeling are presented in online resource 1 (Fig S5 and S6).
Pharmacokinetic parameters for the 1TC model are presented in Table 3. The highest Vt values were found in raphe nuclei (median and dorsal), cingulate, amygdala, and insula. The lowest Vt values were in cerebellum white matter, frontal lobe white matter, and corpus callosum, all tested as reference regions. The cerebellum (grey + white matter), also tested as a reference region, showed intermediate Vt values.
Linear regressions of DVRLREF and DVRSRTM, compared to DVR1TC with the four reference regions tested, are presented in Table 4. For LREF model, all coefficients of determination (R2) were very high (> 0.9). Best fitting was obtained with LREFCERWM and LREFCER (0.95 ± 0.03 both). Note that both LREFCC and LREFFLWM showed high R2 values (0.94 ± 0.03 and 0.94 ± 0.04, respectively). Intercept ranged from 0.01 ± 0.05 (LREFCER) to 0.04 ± 0.08 (LREFCC) thus best fitting (closest to 0) was obtained with LREFCER. Slopes ranged from 0.93 ± 0.02 (LREFCC) to 1.01 ± 0.03 (LREFCERWM) and the best slope value was found with LREFCERWM (1.01 ± 0.02). For the SRTM model, regression parameters (R2) were slightly lower than those found with LREF but close to 0.9, except for CC which showed the lowest R2 value (0.59 ± 0.10). Intercepts ranged from 0.04 ± 0.09 for SRTMCER (best fitting) to 0.68 ± 0.12 for SRTMCC. Slopes ranged from 0.67 ± 0.13 (SRTMCC) to 0.93 ± 0.13 (SRTMCER). SRTMCERWM and SRTMFLWM showed acceptable slopes with 0.92 ± 0.09 and 0.91 ± 0.10, respectively.
Parametric images were obtained using models with reference region. As an example, Fig. 3 shows a brain parametric image of binding potential (BP) using the LREF modeling method with CERWM as reference region.
Test–retest reproducibility and reliability
Test–retest study results are presented in Table 4. Reproducibility was high for the LREF method and satisfying for SRTM. Biases were similar between modeling methods and between reference regions. Results ranged from − 1.48 to 0.95% with the lowest bias for LREFCERWM (-0.04%). As concerns variability (VAR), results were more disparate between methods. The LREF method showed a very low variability of less than 3% for all reference regions. The lowest VAR was found for LREFCEWRM with 2.41%. VAR values for other reference regions were also very low with 2.55%, 2.81%, and 2.84% for LREFFLWM, LREFCER, LREFCC, respectively. SRTM showed higher values. The lowest VAR was SRTMCER with 7.16%. Variability for SRTMCERWM and SRTMFLWM was also satisfying with 8.76% and 8.51%, respectively. Reliability showed better results for the LREF method than with SRTM, with all ICC around 0.9 and the best performance for LREFCERWM (0.95 ± 0.04). The STRM method showed ICC values that were lower and with spread over a wider range from 0.47 ± 0.31 for SRTMFLWM to 0.75 ± 0.26 for SRTMCER. ICCs for SRTMCERWM and SRTMCC were intermediate with 0.63 ± 0.23 and 0.67 ± 0.17, respectively.
Table 5 summarizes reproducibility and reliability parameters in each region for SRTMCERWM and LREFCERWM. DVR was highly correlated between test and retest whatever the method used (R2 > 0.99 for LREFCERWM and R2 > 0.98 for SRTMCERWM). ICC, bias, and VAR per region confirmed the results described in Table 4 and showed that the overall best parameters were those observed using the LREFCERWM method.
Considering LREFCERWM, bias was consistently low, between − 1 and 1%, for all regions. Only the median raphe nucleus showed bias outside this range (− 1.27%). Variability was very low for all regions, with values around or less than 3%, except for the median raphe nucleus (6.62%). Reliability was high (> 0.7) for all regions. Considering SRTMCERWM, biases were all satisfying, with higher values observed for the frontal cortex (− 4.33%) and hippocampus (3.11%). For variability, the median raphe nucleus showed the highest value (20.03%). The temporal (superior and inferior), occipital, parietal cortex, and corpus callosum showed VAR higher than 10%. Other regions showed variability of less than 10%. Reliability was low for the median raphe nucleus (− 0.07), cerebellum (0.33), and cerebellum grey matter (CERGM = 0.35). Some regions showed particularly high reliability with ICC values above 0.7, such as the cingulate (0.74) and frontal cortex (0.86), and the highest ICC value was found for the parahippocampal gyrus (0.95). Other regions showed intermediate ICC from 0.5 to 0.7 such as the hippocampus (0.62) and amygdala (0.65).
Discussion
In this study, we describe the first-in-human trial of a new PET radiopharmaceutical [18F]F13640 which, thanks to its agonist properties, permits in vivo imaging of functional human 5-HT1A receptors. Several analyses were carried out: (i) full kinetic modeling of the radiopharmaceutical using dynamic scans and arterial blood sampling; (ii) comparison of four different reference regions and evaluation of two simplified modeling methods to measure binding potential values and obtain parametric images; and (iii) assessment of test–retest reliability for simplified models. The main finding is that [18F]F13640 constitutes a 5-HT1A receptor radiopharmaceutical with favorable brain binding properties, and the present study describes a PET acquisition protocol and a quantification method suitable for the clinical study of functional 5-HT1A receptors in neurology and psychiatry.
Study design
Initial tests used a protocol with 90 min PET/CT scans and AIF for study participants. However, preliminary analyses of the 5 first subjects revealed a slow washout from cerebral tissues of the radiopharmaceutical, a characteristic that has also been observed for some other radiopharmaceuticals, such as [18F]fallypride [33]. This observation is in accordance with our preclinical observations in rat, cat, and macaque [21]. Since [18F]F13640 has a high affinity for the target sites, the low tissue clearance k2 observed on kinetics could be interpreted as a very low dissociation rate on 5-HT1A receptors. On the basis of these preliminary observations, we concluded that kinetic modeling of the tracer and parameters identification would require a longer acquisition protocol, which was then implemented for the main study. A 165-min long PET acquisition was implemented with a first scan lasting 90 min and a second one lasting 75 min, separated by a 1-h break outside of the camera. This longer protocol with two acquisition periods was facilitated using a hybrid PET-MR scanner, which is capable of brain anatomical realignment and motion correction between PET acquisitions.
Modeling study
Our data revealed that the pharmacokinetic parameters of [18F]F13640 were similar to those found with F13640 (i.e., NLX-112, befiradol) at pharmacological doses. Thus, the fraction of [18F]F13640 parent compound remained high (95%) in blood at 30 to 205 min after injection, consistent with observations on unlabeled NLX-112 in healthy volunteers (Neurolixis, data on file). The ratio of [18F]F13640 binding in plasma to whole blood showed favorable results with a higher distribution in plasma (1.79 ± 0.03). Also, the fp of about 2% was similar between NLX-112 and [18F]F13640. However, we observed unreasonably high K1 values since they were higher than cerebral blood flow investigated with [15O]H2O PET [1, 2]. A plausible, but not a necessarily unique explanation, is that fp measurement has a highly variable value in the first few minutes, and stabilizes later towards a very low value. Processing of the blood samples required 20 to 25 min before being measured for protein binding, and thus may not represent the “true” fp at an early time of acquisition. Therefore, K1 estimates are prone to inaccuracy since fp may not have reached its low equilibrium at short time points after injection. Based on this observation, we also performed 1TC modeling using only mean late-time fp value since they were considered to be more reliable and in accordance with data generated using unlabeled F13640 (Neurolixis, data on file). As expected, this modeling study showed more realistic values (see table S1 in online resource). It should be noted that the calculation of K1 does not constitute an obstacle for use of [18F]F13640 because data generated using this radiopharmaceutical is intended to be analyzed with a simplified method or under static scans with reference region and thus will not require fp value.
A crucial step in the validation of a new brain receptor radiopharmaceutical is the validation of a reference region. Based on literature describing regions poor in 5-HT1A receptors, we tested four different reference regions: corpus callosum (CC), cerebellum (CER), cerebellum white matter (CERWM), and frontal lobe white matter (FLWM). We tested their accuracy and reproducibility performance with two simplified modeling methods, the LREF and SRTM. For the validation with a gold standard, very favorable correlations have been found with the LREF model compared to the 1TC AIF, whatever the reference region. The SRTM method also showed satisfying results in regression whereas compared to 1TC AIF model, except for CC as reference, showed the weakest results. For test–retest reproducibility, ICC, bias, and variability were similar for LREF with the four reference regions, with superior performance when using CERWM as reference region. With SRTM methods, performance for reliability is more mixed, with a clear superiority for the CERWM reference region. Some assumptions need to be validated for the use of SRTM: reference region has no specific binding, the K1/k2 parameter needs to be the same in the reference region, and regions of interest and kinetics can be fitted with the 1TC model. All these assumptions were confirmed with [18F]F13640 since CERWM is known to have very low 5-HT1A receptors expression [35], the K1/k2 parameter was similar between CERWM and regions of interest such as hippocampus or cingulate, and CERWM was fitted with the 1TC model. Taken together, these results indicate that CERWM is the best reference region, showing a low binding, a high correlation with the AIF model, and favorable test–retest reproducibility. Both the LREF and SRTM methods showed similar binding values for all brain regions. Using the SRTM method, the amygdala and hippocampus showed high BP while LREF-calculated BP values were intermediate. According to the 1TC model, the binding values of the amygdala and hippocampus calculated with SRTM are more accurate than the one with LREF. However, considering test–retest parameters, better reproducibility and reliability were found with the LREF method. The high level of test–retest reliability is confirmed at the regional level for LREF with the CERWM, something which is not systematically the case for SRTM and CERWM.
Overall, these results lead us to favor the LREF model with CERWM as reference region for future studies comparing patients to healthy volunteers or patients at different times. The low variability and high reliability (ICC > 0.7) of the LREF model with CERWM as reference region guarantee the power of future studies and the capacity to detect differences between groups due to pathological changes rather than inter-participant or protocol variability. However, the SRTM modeling method and FLWM as an alternative reference region are not to be totally discarded. Indeed, the choice of the modeling method and of the reference region will be driven by the pathology in the study. As an example, cerebellar atrophy could be a confounding factor when using CERWM as a reference region and FLWM would be a suitable alternative. In the present study, the LREF method with CERWM as reference region was used to evaluate [18F]F13640 binding patterns, as discussed below.
Finally, modeling was also computed with the first 90 min (online resource 1 Fig S7). Correlations of Vt by subject, evaluated with 90 min versus 225 min were acceptable but somewhat lower than for shorter scans (R2 between 0.55 to 0.75), with a slight bias (slopes between 0.66 and 1.05) and small intercepts (0.22–0.61). Performing scans of 90 min are therefore feasible but not optimal for an accurate evaluation of the binding potential.
[18F]F13640 brain binding patterns
[18F]F13640 showed elevated BP values in regions known to express a high density of 5-HT1A receptors, including the raphe (median and dorsal) and cortical regions (cingulate, frontal, and temporal superior) [36]. It is noteworthy that the hippocampus only showed intermediate uptake of [18F]F13640 with moderate BP values. Median and dorsal raphe nuclei were regions with the highest BP values but also with the most substantial variability. This can be explained by the fact that the raphe nucleus is the only region that is not defined on the basis of anatomical data but by functional data [29]. Note that some vessels showed also high binding value and could affect the BP of the nearest regions due to a partial volume effect.
[18F]F13640 binding in cerebellum grey matter
As described above, cerebellum grey matter and vermis both showed high BP values. Whereas a high expression of 5-HT1A receptors has been described in vermis [35, 37], it is frequently postulated that cerebellum grey matter does not express 5-HT1A receptors. Indeed, according to post-mortem autoradiographic studies [38, 39], 5-HT1A receptors are expressed in fetal and neonatal stages but not in adults [40]. The present unexpected binding of [18F]F13640 in the cerebellum could thus question its target specificity, but an interaction by the radiotracer with a possible off-target seems unlikely based on well-documented in vitro studies on F13640 [20]. Moreover, our previous preclinical studies also detected [18F]F13640 binding in the cerebellum in different species [21, 22] and, in all these studies, co-administration of pharmacologically relevant doses of a 5-HT1A receptor agonist (8-OH-DPAT) or of an antagonist (WAY-100635) inhibited [18F]F13640 binding, indicating that its binding in cerebellum grey matter specifically reflects 5-HT1A receptor expression in this region. In addition to the present data with [18F]F13640, the assumption of a total lack of 5-HT1A receptors in the cerebellum was already challenged by previous PET studies revealing [carbonyl-11C]WAY-100635 binding in human cerebellum [35], with more pronounced labeling in grey matter [37]. In other studies, participants were excluded due to cerebellar TACs outside the range of the control population when the cerebellum was used as a reference region [41,42,43]. Other 5-HT1A receptor radiopharmaceuticals also showed marked binding in the cerebellum, notably [11C]CUMI-101 which showed a significant reduction of labeling following 8-OH-DPAT or WAY-100635 pre-injection [44]. Taken together, these observations indicate that 5-HT1A receptors are indeed present in cerebellum grey matter, excluding the cerebellum (which contain the grey matter) as a reference region for [18F]F13640.
[18F]F13640 binding in other brain regions
In comparison with other 5-HT1A radiopharmaceuticals, [18F]F13640 showed particularly high uptake in raphe nuclei, cortical regions, and vermis in limbic and paralimbic regions. Although [18F]MPPF also showed high uptake in raphe nuclei, it preferentially binds to limbic areas such as the hippocampus and amygdala whereas cortical regions showed less uptake [45]. Another antagonist radiopharmaceutical, [carbonyl-11C]WAY-100635, also showed high binding in the raphe and frontal cortex (like [18F]F13640) but it also showed marked binding to the hippocampus (like [18F]MPPF) [3]. Binding differences between [carbonyl-11C]WAY-100635, [18F]MPPF and [18F]F13640 are likely to be due to pharmacological differences between these 5-HT1A receptor radiopharmaceuticals [46, 47]. We hypothesize that some regions such as the hippocampus, which showed high uptake with antagonist radiopharmaceuticals and less with the agonist [18F]F13640, express a high proportion of G-protein uncoupled 5-HT1A receptors. In contrast, regions with higher [18F]F13640 binding values, such as the cingulate cortex, frontal cortex, or vermis, may express a relatively higher proportion of functionally active, G-protein-coupled receptors. Finally, regions such as raphe which showed high binding with both antagonist and agonist radiopharmaceuticals may express both states of 5-HT1A receptors. Thus, differences in receptor binding patterns can be found between agonists and antagonists radiopharmaceuticals targeting the same receptor. An in vitro study using [18F]F13640 and [18F]MPPF already explored this hypothesis and showed differences in binding between agonists and antagonists radiotracer with 5-HT1A in Alzheimer’s disease [48]. Analogous results were found with D2/3 dopamine receptors, for which the agonist, [3H]NPA, showed a greater extent of than the antagonist [11C]raclopride binding in a rat Parkinson’s disease model [49].
Perspectives opened by [18F]F13640
Based on the above observations, it can be considered that [18F]F13640 has now the status of a radiopharmaceutical whose novel pharmacokinetic and radiopharmacological characteristics offer new perspectives in neurology and psychiatry.
First of all, the sustained binding of [18F]F13640 will allow studies to be performed with multiple acquisitions at different late time points following the same injection. This experimental paradigm could, for example, allow exploration of changes in receptor coupling states associated with circadian rhythms. As an illustration, a previous preclinical study demonstrated that [18F]F13640 is sensitive to fluctuations in serotonin levels [22] opening the way for exploration of in vivo serotonin release in physiological or pathological processes.
As demonstrated in the present study, [18F]F13640 binding using the LREF method showed excellent test–retest parameters, a profile which makes it attractive for studies involving repeated measurements on the same subject, including drug occupancy and intervention studies, or for exploring pathological states at different time points—situations where reproducibility and reliability of the PET measurements is crucial.
In terms of its radiopharmacological characteristics, the binding pattern of [18F]F13640 opens the way to informative imaging studies in neurology and psychiatry. For example, [18F]F13640 shows high binding values in the raphe, consistent with the therapeutic-like activity of 5-HT1A receptor agonists in rodent and non-human primate models of L-DOPA-induced dyskinesia (LID) in Parkinson’s disease [50,51,52] which is the focus of an ongoing clinical study (ClinicalTrials.gov Identifier: NCT05148884). [18F]F13640 also shows high binding values in cortical regions associated with antidepressant activity or control of negative symptoms of schizophrenia [1,2,3]. 5-HT1A receptors in the brainstem are also promising targets to alleviate respiratory dysfunction in disorders such as Rett syndrome [56]. [18F]F13640 could therefore be a promising tool to assess the role of 5-HT1A receptors in a variety of different disorders involving serotonergic mechanisms [15].
Conclusion
The present study reports the first-in-human validation and full kinetic modeling of [18F]F13640 as the first 5-HT1A receptor agonist usable as a PET radiopharmaceutical. [18F]F13640 shows many favorable radiopharmacological and radiopharmaceutical characteristics: radiolabeling with fluorine-18, high selectivity over cross-reacting sites, high reproducibility, and long-term binding which facilitates the experimental protocols. [18F]F13640 shows pronounced binding in raphe nuclei and cortical regions, with notable differences in comparison with classical antagonist PET radiopharmaceuticals. All these characteristics confirm the interest of developing an agonist radiotracer able to specifically target functionally active 5-HT1A receptors in studies with long-term scans and test–retest protocols in order to investigate disease states in neurology and psychiatry.
Data Availability
The datasets generated during this study are available from the corresponding author.
References
Polter AM, Li X. 5-HT1A receptor-regulated signal transduction pathways in brain. Cell Signal. 2010;22:1406–12.
Pike VW, McCarron JA, Lammerstma AA, Hume SP, Poole K, Grasby PM, et al. First delineation of 5-HT1A receptors in human brain with PET and [11C]WAY-100635. Eur J Pharmacol. 1995;283:R1–3.
Gunn RN, Sargent PA, Bench CJ, Rabiner EA, Osman S, Pike VW, et al. Tracer kinetic modeling of the 5-HT1A receptor ligand [carbonyl-11C]WAY-100635 for PET. NeuroImage. 1998;8:426–40.
Zhuang Z-P, Kung M-P, Kung HF. Synthesis And Evaluation of 4-(2’-Methoxyphenyl)-1-[2’-[N-(2”-pyridinyl)- p-iodobenzamido]ethyl]piperazine (p-MPPI): A new iodinated 5-HT1A ligand. J Med Chem. 1994;37:1406–7.
Passchier J, van Waarde A, Pieterman RM, Elsinga PH, Pruim J, Hendrikse HN, et al. Quantitative imaging of 5-HT(1A) receptor binding in healthy volunteers with [(18)f]p-MPPF. Nucl Med Biol. 2000;27:473–6.
Passchier J, van Waarde A, Pieterman RM, Elsinga PH, Pruim J, Hendrikse HN, et al. In vivo delineation of 5-HT1A receptors in human brain with [18F]MPPF. J Nucl Med. 2000;41:1830–5.
Costes N, Merlet I, Ostrowsky K, Faillenot I, Lavenne F, Zimmer L, et al. A 18F-MPPF PET normative database of 5-HT1A receptor binding in men and women over aging. J Nucl Med. 2005;46:1980.
Merlet I, Ostrowsky K, Costes N, Ryvlin P, Isnard J, Faillenot I, et al. 5-HT1A receptor binding and intracerebral activity in temporal lobe epilepsy: an [18F]MPPF-PET study. Brain. 2004;127:900–13.
Derry C, Benjamin C, Bladin P, le Bars D, Tochon-Danguy H, Berkovic SF, et al. Increased serotonin receptor availability in human sleep: evidence from an [18F]MPPF PET study in narcolepsy. Neuroimage. 2006;30:341–8.
Kepe V, Barrio JR, Huang S-C, Ercoli L, Siddarth P, Shoghi-Jadid K, et al. Serotonin 1A receptors in the living brain of Alzheimer’s disease patients. Proc Natl Acad Sci U S A. 2006;103:702–7.
Meyer M, Lamare F, Asselineau J, Foubert-Samier A, Mazère J, Zanotti-Fregonara P, et al. Brain 5-HT1A receptor binding in multiple system atrophy: An [18 F]-MPPF PET study. Mov Disord. 2021;36:246–51.
Billard T, Bars D, Zimmer L. PET radiotracers for molecular imaging of serotonin 5-HT1A receptors. CMC. 2013;21:70–81.
Gozlan H, Thibault S, Laporte AM, Lima L, Hamon M. The selective 5-HT1A antagonist radioligand [3H]WAY 100635 labels both G-protein-coupled and free 5-HT1A receptors in rat brain membranes. Eur J Pharmacol. 1995;288:173–86.
Watson J, Collin L, Ho M, Riley G, Scott C, Selkirk JV, et al. 5-HT(1A) receptor agonist-antagonist binding affinity difference as a measure of intrinsic activity in recombinant and native tissue systems. Br J Pharmacol. 2000;130:1108–14.
Newman-Tancredi A, Depoortère RY, Kleven MS, Kołaczkowski M, Zimmer L. Translating biased agonists from molecules to medications: Serotonin 5-HT1A receptor functional selectivity for CNS disorders. Pharmacol Ther. 2021;107937.
Hendry N, Christie I, Rabiner EA, Laruelle M, Watson J. In vitro assessment of the agonist properties of the novel 5-HT1A receptor ligand, CUMI-101 (MMP), in rat brain tissue. Nucl Med Biol. 2011;38:273–7.
Shrestha SS, Liow J-S, Lu S, Jenko K, Gladding RL, Svenningsson P, et al. 11 C-CUMI-101, a PET radio ligand, behaves as a serotonin 1A receptor antagonist and also binds to α 1 adrenoceptors in brain. J Nucl Med. 2014;55:141–6.
Selvaraj S, Turkheimer F, Rosso L, Faulkner P, Mouchlianitis E, Roiser JP, et al. Measuring endogenous changes in serotonergic neurotransmission in humans: a [11C]CUMI-101 PET challenge study. Mol Psychiatry. 2012;17:1254–60.
Girgis RR, Forbes A, Abi-Dargham A, Slifstein M. A positron emission tomography occupancy study of brexpiprazole at dopamine D2 and D3 and serotonin 5-HT1A and 5-HT2A receptors, and serotonin reuptake transporters in subjects with schizophrenia. Neuropsychopharmacology. 2020;45:786–92.
Colpaert FC, Tarayre JP, Koek W, Pauwels PJ, Bardin L, Xu X-J, et al. Large-amplitude 5-HT1A receptor activation: a new mechanism of profound, central analgesia. Neuropharmacology. 2002;43:945–58.
Vidal B, Fieux S, Colom M, Billard T, Bouillot C, Barret O, et al. 18F-F13640 preclinical evaluation in rodent, cat and primate as a 5-HT1A receptor agonist for PET neuroimaging. Brain Struct Funct. 2018;223:2973–88.
Colom M, Vidal B, Fieux S, Redoute J, Costes N, Lavenne F, et al. [18F]F13640, a 5-HT1A receptor radiopharmaceutical sensitive to brain serotonin fluctuations. Front Neurosci. 2021;15:622423.
Mérida I, Reilhac A, Redouté J, Heckemann RA, Costes N, Hammers A. Multi-atlas attenuation correction supports full quantification of static and dynamic brain PET data in PET-MR. Phys Med Biol. 2017;62:2834–58.
Reilhac A, Merida I, Irace Z, Stephenson MC, Weekes AA, Chen C, et al. Development of a dedicated rebinner with rigid motion correction for the mMR PET/MR scanner, and validation in a large cohort of 11 C-PIB scans. J Nucl Med. 2018;59:1761–7.
Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. NeuroImage. 2010;51:221–7.
Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19:224–47.
Gousias IS, Rueckert D, Heckemann RA, Dyet LE, Boardman JP, Edwards AD, et al. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. NeuroImage. 2008;40:672–84.
Faillenot I, Heckemann RA, Frot M, Hammers A. Macroanatomy and 3D probabilistic atlas of the human insula. NeuroImage. 2017;150:88–98.
Mottolese R, Redouté J, Costes N, Le Bars D, Sirigu A. Switching brain serotonin with oxytocin. Proc Natl Acad Sci USA. 2014;111:8637–42.
Logan J. Graphical analysis of PET data applied to reversible and irreversible tracers. Nuclear Medicine and Biology. 2000;27:661–70.
Gunn RN, Lammertsma AA, Hume SP, Cunningham VJ. Parametric Imaging of Ligand-Receptor Binding in PET Using a Simplified Reference Region Model. NeuroImage. 1997;6:279–87.
Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86:420–8.
Kegeles LS, Slifstein M, Xu X, Urban N, Thompson JL, Moadel T, et al. Striatal and extrastriatal dopamine D2/D3 receptors in schizophrenia evaluated with [18F]fallypride positron emission tomography. Biol Psychiatry. 2010;68:634–41.
Vestergaard MB, Calvo OP, Hansen AE, Rosenbaum S, Larsson HBW, Henriksen OM, et al. Validation of kinetic modeling of [15O]H2O PET using an image derived input function on hybrid PET/MRI. NeuroImage. 2021;233:117950.
Ohta S, Meyer E, Fujita H, Reutens DC, Evans A, Gjedde A. Cerebral [ 15 O]water clearance in humans determined by PET: I. theory and normal values. J Cereb Blood Flow Metab. 1996;16:765–80.
Parsey RV, Arango V, Olvet DM, Oquendo MA, Van Heertum RL, Mann JJ. Regional heterogeneity of 5-HT 1A receptors in human cerebellum as assessed by positron emission tomography. J Cereb Blood Flow Metab. 2005;25:785–93.
Barnes NM, Sharp T. A review of central 5-HT receptors and their function. Neuropharmacology. 1999;38:1083–152.
Hirvonen J, Kajander J, Allonen T, Oikonen V, Någren K, Hietala J. Measurement of serotonin 5-HT1A receptor binding using positron emission tomography and [ carbonyl - 11 C]WAY-100635—considerations on the validity of cerebellum as a reference region. J Cereb Blood Flow Metab. 2007;27:185–95.
Hall H, Lundkvist C, Halldin C, Farde L, Pike VW, McCarron JA, et al. Autoradiographic localization of 5-HT1A receptors in the post-mortem human brain using [3H]WAY-100635 and [11C]way-100635. Brain Res. 1997;745:96–108.
Pazos A, Probst A, Palacios JM. Serotonin receptors in the human brain—III. Autoradiographic mapping of serotonin-1 receptors. Neuroscience. 1987;21:97–122.
del Olmo E, Díaz A, Guirao-Piñeyro M, del Arco C, Pascual J, Pazos A. Transient localization of 5-HT1A receptors in human cerebellum during development. Neurosci Lett. 1994;166:149–52.
Doder M, Rabiner EA, Turjanski N, Lees AJ, Brooks DJ. Tremor in Parkinson’s disease and serotonergic dysfunction: An 11 C-WAY 100635 PET study. Neurology. 2003;60:601–5.
Turner MR, Rabiner EA, Hammers A, Al-Chalabi A, Grasby PM, Shaw CE, et al. [11C]-WAY100635 PET demonstrates marked 5-HT1A receptor changes in sporadic ALS. Brain. 2005;128:896–905.
Rabiner EA, Messa C, Sargent PA, Husted-Kjaer K, Montgomery A, Lawrence AD, et al. A database of [11C]WAY-100635 binding to 5-HT1A receptors in normal male volunteers: normative data and relationship to methodological, demographic, physiological, and behavioral variables. NeuroImage. 2002;15:620–32.
Milak MS, Severance AJ, Ogden RT, Prabhakaran J, Kumar JSD, Majo VJ, et al. Modeling considerations for 11 C-CUMI-101, an agonist radiotracer for imaging serotonin 1A receptor in vivo with PET. J Nucl Med. 2008;49:587–96.
Costes N, Merlet I, Zimmer L, Lavenne F, Cinotti L, Delforge J, et al. Modeling [18 F]MPPF positron emission tomography kinetics for the determination of 5-hydroxytryptamine(1A) receptor concentration with multi injection. J Cereb Blood Flow Metab. 2002;22:753–65.
Kenakin T. Functional selectivity through protean and biased agonism: who steers the ship? Mol Pharmacol. 2007;72:1393–401.
Perez DM, Karnik SS. Multiple signaling states of G-protein-coupled receptors. Pharmacol Rev. 2005;57:147–61.
Vidal B, Sebti J, Verdurand M, Fieux S, Billard T, Streichenberger N, et al. Agonist and antagonist bind differently to 5-HT 1A receptors during Alzheimer’s disease: A post-mortem study with PET radiopharmaceuticals. Neuropharmacology. 2016;109:88–95.
Palner M, Kjaerby C, Knudsen GM, Cumming P. Effects of unilateral 6-OHDA lesions on [3H]-N-propylnorapomorphine binding in striatum ex vivo and vulnerability to amphetamine-evoked dopamine release in rat. Neurochem Int. 2011;58:243–7.
Iderberg H, McCreary AC, Varney MA, Kleven MS, Koek W, Bardin L, et al. NLX-112, a novel 5-HT 1A receptor agonist for the treatment of l -DOPA-induced dyskinesia: Behavioral and neurochemical profile in rat. Exp Neurol. 2015;271:335–50.
Depoortere R, Johnston TH, Fox SH, Brotchie JM, Newman-Tancredi A. The selective 5-HT1A receptor agonist, NLX-112, exerts anti-dyskinetic effects in MPTP-treated macaques. Parkinsonism & Related Disorders. 2020;78:151–7.
Fisher R, Hikima A, Morris R, Jackson MJ, Rose S, Varney MA, et al. The selective 5-HT1A receptor agonist, NLX-112, exerts anti-dyskinetic and anti-parkinsonian-like effects in MPTP-treated marmosets. Neuropharmacology. 2020;167:107997.
Buckley PF. Neuroimaging of schizophrenia: structural abnormalities and pathophysiological implications. Neuropsychiatr Dis Treat. 2005;1:193–204.
Razakarivony O, Newman-Tancredi A, Zimmer L. Towards in vivo imaging of functionally active 5-HT1A receptors in schizophrenia: concepts and challenges. Transl Psychiatry. 2021;11:22.
Newman-Tancredi A, Bardin L, Auclair A, Colpaert F, Depoortère R, Varney MA. NLX-112, a highly selective 5-HT 1A receptor agonist, mediates analgesia and antidepressant-like activity in rats via spinal cord and prefrontal cortex 5-HT 1A receptors, respectively. Brain Res. 2018;1688:1–7.
Abdala AP, Bissonnette JM, Newman-Tancredi A. Pinpointing brainstem mechanisms responsible for autonomic dysfunction in Rett syndrome: therapeutic perspectives for 5-HT1A agonists. Front Physiol [Internet]. 2014 [cited 2021 Nov 24];5. Available from: http://journal.frontiersin.org/article/10.3389/fphys.2014.00205/abstract.
Acknowledgements
This work was supported by AVIESAN-ITMO Technologies for health, by the French Association for Research on Down Syndrome (AFRT), by the Hospices Civils de Lyon, by the imaging platform CERMEP, and by the LABEX PRIMES of Université de Lyon. PC was awarded a scholarship for the French Fund for Medical Research FRM (FDM202006011252).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
AN-T is a shareholder and an employee of Neurolixis. The other authors have no disclosures.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Neurology.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Courault, P., Lancelot, S., Costes, N. et al. [18F]F13640: a selective agonist PET radiopharmaceutical for imaging functional 5-HT1A receptors in humans. Eur J Nucl Med Mol Imaging 50, 1651–1664 (2023). https://doi.org/10.1007/s00259-022-06103-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-022-06103-1






