Abstract
In this paper, we present the novel Deep-MEG approach in which image-based representations of magnetoencephalography (MEG) data are combined with ensemble classifiers based on deep convolutional neural networks. For the scope of predicting the early signs of Alzheimer’s disease (AD), functional connectivity (FC) measures between the brain bio-magnetic signals originated from spatially separated brain regions are used as MEG data representations for the analysis. After stacking the FC indicators relative to different frequency bands into multiple images, a deep transfer learning model is used to extract different sets of deep features and to derive improved classification ensembles. The proposed Deep-MEG architectures were tested on a set of resting-state MEG recordings and their corresponding magnetic resonance imaging scans, from a longitudinal study involving 87 subjects. Accuracy values of 89% and 87% were obtained, respectively, for the early prediction of AD conversion in a sample of 54 mild cognitive impairment subjects and in a sample of 87 subjects, including 33 healthy controls. These results indicate that the proposed Deep-MEG approach is a powerful tool for detecting early alterations in the spectral–temporal connectivity profiles and in their spatial relationships.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Deep convolutional neural networks (CNNs) have become very popular in recent years thanks to their ability to decode images [1], video streams [2], and other biomedical signals [3], including 2D and 3D neuroimaging data [4]. The use of CNNs, in fact, offers the possibility to recognize the presence of patterns that other techniques are not able to reveal. In clinical scenarios, when data availability is limited, transfer learning (TL) can be applied to transfer the knowledge, previously learned by the CNN, for solving new problems faster or with different learning solutions [5, 6]. By combining the merit of multiple classifiers, ensemble learning can be an additional powerful instrument to improve the performance of predictive models [7,8,9,10].
Thanks to these advantages, the use of CNNs is becoming predominant for the analysis of many types of biomedical images, in particular for decoding encephalographic signals, in which it is important to recognize the various neural activation patterns in relation to diseases. Many studies [11,12,13,14] have used deep learning to 1D electroencephalography (EEG) signals to reach this goal, looking for solutions and architectures of different neural networks in order to extract the most discriminating features.
In recent years, the analysis of brain activity has been extended to an innovative technique, known as magnetoencephalography (MEG) [15,16,17,18]. MEG is a powerful non-invasive diagnostic tool that possesses the unique advantage of providing a direct measure of the neural activity of the pyramidal neurons in the brain, ensuring high spatial and temporal resolutions (of order of mm and ms, respectively), and a fast preparation time [18]. A set of MEG recordings, together with the positions of the corresponding sources, encompass complex high-dimensional information on the brain network functioning, which can be difficult to uncover via standard methodologies, as for the case of the Alzheimer’s disease (AD).
AD is a neurodegenerative disorder and the most common form of dementia worldwide [19]. AD may start decades before the symptoms occur and then gradually evolve, with progressive alteration of cognitive and functional abilities. A precursory condition to AD, named mild cognitive impairment (MCI), is known to indicate a deviation from normal aging and an increased risk of developing dementia in future [20]. MCI, which can be caused by disorders other than AD (such as frontotemporal dementia), can remain a stable condition over time (stable MCI or sMCI), or finally progress to AD (progressive MCI or pMCI). The full-blown AD is a disabling condition resulting from the synaptic disruption of local and large-scale networks of the brain for which there is no cure. Finding new methods to detect pre-symptomatic or prodromal phases, i.e., pMCI, and predict earlier their progression toward AD would facilitate the timely implementation of therapeutic strategies [26].
To date, the most effective approaches for early AD diagnosis involve the use of invasive techniques such as the cerebrospinal fluid analysis [21] or the positron emission tomography (PET) [22, 23], which require performing a lumbar puncture or the use of radioactive tracers, respectively. Non-invasive diagnostic tools are being explored as alternatives [24,25,26,27,28], with MEG representing a promising technique to be taken into account [15,16,17,18]. In the following section, we report an overview of the state-of-the-art methods relative to the analysis of MEG data, with particular reference to the early AD diagnosis.
1.1 Literature review
Research on deep learning-based analysis of MEG signals is in progress. Deep learning architectures have been applied for artifacts removal [29] or to decode the brain responses to a set of visual, auditory and somatosensory stimuli [30]. In particular, Croce and colleagues [29] derived spectra and 2D topographic representations of the independent components (IC) of EEG and MEG recordings. The set of ICs was used as input to the convolutional layers of a CNN for the automatic identification of artifacts. The obtained accuracy values outperformed the state-of-the-art feature-based methods for artifact removal. Zubrarev et al. [30] used a mixture of k-latent sources based on a linear autoregressive model to represent the MEG time courses. The authors designed two variants of CNNs, 1D and 2D, to process the temporal dynamics of the obtained signals and applied them to decode the brain responses to a set of visual, auditory, and somatosensory stimuli. Recently, Aoe and colleagues [31] proposed a deep neural network, Mnet, which is based on the EnvNet-v2 [32], an architecture originally designed to classify environmental sounds. By directly analyzing 160 channels of raw MEG signal and the relative powers of six frequency bands, the proposed approach achieved high level of accuracy in the computer-aided diagnosis of spinal cord injury and epilepsy.
Recent studies addressed the discrimination of mild forms of cognitive impairment from healthy subjects. A shallow neural network was used by Amezquita-Sanchez et al. [33] to distinguish 18 MCI patients and 19 control subjects. MEG frequency sub-bands were characterized via ensemble empirical mode decomposition and permutation entropy measures and then classified via an enhanced probabilistic neural network (EPNN). In the work by Lopez-Martin et al. [34], CNN models were used to decode a large set of randomized features, i.e., mean, median, standard deviation, mean absolute deviation, and range, relative to the mutual information between paired MEG time series and rearranged as 2D matrices. Their method outperformed the classic machine learning approaches in the classification of patients as MCI or healthy subjects.
A very promising tool for the neuroimaging research community is represented by the MEG-based measures of functional connectivity (FC) [15,16,17, 35,36,37,38]. FC analysis can be performed in relation to different frequency bands, and it is capable of providing a huge amount of information on the relationship between the brain regions and on their organization into large-scale networks. In fact, a reduced [36, 37] or increased [15,16,17] synchronization between the activities of key brain regions has been revealed in AD patients by means of FC, posing MEG-based FC as a promising biomarker to evaluate AD progression. The bio-magnetic activity of AD patients, from a spectral perspective, is generally associated with changes in the θ, β, and α bands [39]. Similar patterns have been also observed in the more severe forms of MCI [39], suggesting that MEG-based spectral characteristics are fundamental indices for AD diagnosis. The β band oscillations, in particular, have been proposed as quantitative indicators to predict the progression to AD at the MCI stage [40,41,42,43], while higher synchronization in low-frequency bands, e.g., the θ band, has been observed in MCI groups as compared to the control healthy groups [39, 44].
Recently, based on the analysis of the functional strength between pre- and post-conversion MEG scans, Pusil and colleagues [38] succeeded in automatically detecting all the MCI subjects progressing to AD. Their analysis was based on the multivariate connectivity phase estimation (PCE) in five MEG frequency bands using both pre- and post-conversion MEG data. However, the temporal, power spectrum, and the topological properties of MEG data seem to drive complementary information [17] that can be further characterized and investigated to detect AD earlier and before the symptoms occur. In fact, the diagnostic prediction of the conversion of MCI to AD, using MEG data of the asymptomatic at-risk stage, i.e., not showing clinical evidence of AD, is still an open problem [15,16,17, 36, 37]. To our knowledge, neither deep CNNs nor ensemble architectures have been deployed to recognize the FC alterations due to the early phases of AD. We believe that deep learning can help decoding the more subtle changes in the brain network activity occurring during the early phases of AD progression, increasing the predictive capability of the automated approaches to the analysis of MEG data [45, 46].
1.2 The proposed method
In this work, we propose to exploit transfer learning via pre-trained CNNs to decode FC maps, whose explicitness to human eye is not trivial, with the aim to reveal the topography of the neural activations based on MEG/MRI data. Indeed, the key to uncover early signs of AD may be hidden in the multidimensional nature of FC maps which encode not only information on the electrical coupling between spatially distant neuronal populations but also on the way such neuronal activity is spatially distributed and coordinated [15,16,17].
As a continuation of our preliminary study [47], in the present work we take a step further in decoding the electrophysiological anomalies occurring before the conversion to AD (early diagnosis) by proposing a deep learning approach, named Deep-MEG, which exploits the new paradigm image-based coding of MEG/MRI data, and different ensemble classification architectures. The proposed methods include:
-
(1)
The extraction of temporal, multi-frequency, and spatial data from MEG recordings and MRI scan in the form of FC maps;
-
(2)
The novel coding of the FC maps into deep features by using transfer learning;
-
(3)
The implementation of an ensemble learning architecture to cooperatively combine the decision of multiple predictive modules on the basis of different FC mapping.
Deep-MEG differs from the other existing approaches in the way the FC patterns of the brain network are decoded by 2D CNNs. Being the FC maps used in our approach topologically organized based on the subject-specific MEG/MRI source reconstruction, Deep-MEG derives information not only on the individual hypo- or hyper-synchronization responses, but also on the 2D patterns related to the spatial arrangement of FC values within the maps, corresponding to an information route on the brain connectome up to now unexplored [48]. Pre-trained deep CNNs provide us the means to decode those FC patterns via transfer learning, i.e., without the need for large datasets to set the network parameters at the training level. Ensemble classifiers, in addition, led us to extend our analysis to selected frequency bands, stressing the role of definite spectral profiles activities that are associated with different levels of AD progression [39,40,41,42,43,44].
To evaluate the predictive performance of the proposed system, we performed quantitative experiments on data from a longitudinal study from the Hospital Universitario San Carlos (Madrid, Spain) [15], involving 54 MCI patients (of which 27 pMCI patients who progressed toward AD during a 3-year follow-up) and 33 healthy controls (HC).
The rest of the paper is organized as follows. In Sect. 2, we describe the characteristics of the subjects involved in the study and the acquisition process of MEG recordings and MRI scans. In Sect. 3, we describe the methods used in our study. The preprocessing of spatial and temporal data with the band-filtering operations, the different variants of FC indicators and their mapping into FC images, the derivation of deep spatiotemporal features based on AlexNet [49], and the ensemble learning architectures for the two-class and three-class scenarios [50]. Experimental results obtained with the proposed pipeline for investigation and comparison with other existing approaches are reported in Sect. 4. Finally, a discussion of the results is included in Sect. 5.
2 Materials
2.1 The case study
A total of 54 MCI patients were recruited from the Hospital Universitario San Carlos (Madrid, Spain) [15], and 33 healthy controls were enrolled in this study after signing an informed consent. In Table 1, we present the demographic characteristics of the participants. All of them were right-handed [51]. The study was approved by the Hospital Universitario San Carlos Ethics Committee (Madrid). A diagnosis of MCI was made on 54 patients according to the National Institute on Aging-Alzheimer Association (NIA-AA) clinical criteria [52]. Additional 33 elderly healthy subjects were included in the present work as control (HC). Besides meeting the clinical criteria, MCI participants had signs of neuronal injury (hippocampal volume measured by MRI). Thus, they might be considered as “MCI due to AD” with an intermediate likelihood [52]. The MCI patients were cognitively and clinically followed up for approximately 3 years (every six months) and were split into two groups, i.e., sMCI and pMCI, according to their clinical outcome. The sMCI group (n = 27) was comprised of those participants that still fulfilled the diagnosis criteria of MCI at the end of follow-up. The pMCI group (n = 27) was composed of those subjects that met the criteria for probable AD at the end of the follow-up [53]. None of the participants had a history of psychiatric or neurological disorders (other than MCI or AD). General inclusion criteria were: age between 65 and 80, a modified Hachinski score [54] ≤ 4, a short-form Geriatric Depression Scale score ≤ 5, and T1 MRI within 12 months and 2 weeks before the two MEG recordings without indication of infection, infarction, or focal lesions (rated by two independent experienced radiologists) [55]. Patients were off those medications that could affect MEG activity, such as cholinesterase inhibitors, 48 h before recordings.
2.2 MRI acquisitions
3D T1-weighted anatomical brain magnetic resonance imaging (MRI) scans were collected with a General Electric 1.5 T MRI scanner, using a high-resolution antenna and a homogenization PURE filter (Fast Spoiled Gradient Echo, FSPGR, sequence with parameters: TR/TE/TI = 11.2/4.2/450 ms; flip angle 12°; 1 mm slice thickness; a 256 × 256 matrix; and FOV 25 cm).
2.3 MEG recordings
Weighted MEG recordings were acquired with a 306-channel Vectorview system (Elekta Neuromag) at the Center for Biomedical Technology (Madrid, Spain). MEG recordings were collected at the same time of the day in two different periods:
-
(1)
pre-conversion stage (54 MCIs and 33 HCs), at baseline (first MEG);
-
(2)
post-conversion stage (27 sMCIs and 27 pMCIs), 24 ± 6 months after the first MEG (second MEG).
In both the sets of MEG recordings, participants were in an awake, resting state with their eyes closed. For each subject, 5-min task-free data were recorded at a sampling frequency of 1000 Hz. In the present study, the baseline pre-conversion data are used to test the predictive power of the system with reference to the early signs of AD in pMCI subjects, i.e., when the dementia is still not present. Post-conversion data in which signs of AD are clinically evident in pMCI subjects but not in sMCI subjects are used for comparative analysis.
3 Methods
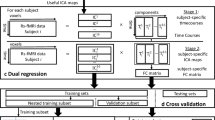
A schematic representation of the pipeline of the proposed platform is given in Fig. 1. The main characteristics of the methods are as follows:
-
i.
MEG recordings and MRI scan are processed to derive temporal, multi-frequency, and spatial data. The system receives as input a set of MEG recordings and corresponding MRI scan. Sensor-space MEG signals are filtered in different frequency. The MRI scan is used to reconstruct the MEG signal at the neural sources and derive the spatial relationships among the measured MEG time series [56,57,58]. The statistical interdependence between MEG signals measured at two or more spatially separated brain regions is quantified through functional connectivity (FC) indices [35], obtained as measures of phase or envelope synchronization for different frequency bands.
-
ii.
FC is coded into image-based representations. The intricate communication patterns among neuronal populations are represented by the spatial arrangements of pixel values in a set of FC images. Each image is generated by mapping the variants of FC indices into a topologically organized 2D space. Different FC indicators or indicators of sub-band frequencies are also combined into RGB images to acquire a new set of deep CNN features that can boost the classification performance. Hyper- or hypo-activation patterns in the FC images reflect both the topological organization and the functioning of the brain network.
-
iii.
Deep-MEG features are used to decode the FC patterns. The information patterns in each generated FC image, which represent the spatiotemporal interdependence of signaling in the brain network, are hierarchically decomposed by means of the CNN layers of AlexNet [49], which is used as baseline architecture for deep features extraction. Relevant features are automatically selected in relation to the classification task. The obtained deep spatiotemporal features allow a new representation of the intricate structure of MEG-based FC that standard engineered features may fail to extract.
-
iv.
An ensemble learning architecture combines the decision of multiple predictive modules. The obtained variants of FC images, also relative to different frequency bands, are used to train a set of base-predictive modules with linear discriminant analysis (LDA) or support vector machine (SVM) classifiers [50]. Each base classifier receives the relevant deep features automatically selected from a single FC image. The predictive scores of the base classifiers are then combined to derive the final assignment.
3.1 Artifacts removal, segmentation, and band filtering
MEG recordings were first band-pass-filtered online between 0.1 and 330 Hz. Then, the Maxfilter software (Elekta Neuromag® v2.2, correlation threshold = 0.9, time window = 10 s) was used to remove external noise of the raw MEG data with the temporal extension of the signal space separation method with movement compensation [59]. MEG data were automatically scanned for ocular, muscle, and jump artifacts using the Fieldtrip software [56]. Subsequently, artifacts were visually confirmed and removed by a MEG expert. The remaining artifact-free data were segmented in 4 s segments (epochs), as shown in Fig. 1. An independent component analysis-based procedure was used to remove the heart magnetic field artifact. Previously to source data calculation, MEG signals were filtered into θ (4–8 Hz), α (8–12 Hz), β (12–30 Hz), and γ (30–55 Hz) frequency bands with a 1800-order finite impulse response filter with Hamming window and a two-pass filtering procedure. Being the beta band very wide, for some analyses it was useful to further divide it into β1 (12–20 Hz) and β2 (20–30 Hz).
3.2 Source reconstruction and brain parcellation
We employed Freesurfer software (version 5.1.0.21) [60] to obtain the cortex, skull, and scalp segmentation. A regular grid with 10-mm spacing was created in the brain template from the Montreal Neurological Institute (MNI). This set of nodes was transformed to each participant’s space using a nonlinear normalization between the native T1 image (whose coordinate system was previously converted to match the MEG coordinate system) and a standard T1 in the MNI space. The forward model was solved with a single-shell method [61] with a unique boundary defined by the inner skull (the combination of white matter, gray matter, and cerebrospinal fluid) taken from the individual T1. We carried out the source reconstruction independently for each subject and frequency band, using a linearly constrained minimum variance (LCMV) beamformer [62]. Beamforming filters were estimated with normalized lead fields, regularized covariance matrices averaged over trials, and a 1% regularization factor (Fig. 1). The neural MEG sources so derived were anatomically parcellated by dividing the cortex into 90 regions of interest (ROIs) according to the AAL atlas [58] as shown in Fig. 1.
3.3 Functional connectivity analysis
The spatial, temporal, and band-filtered data extracted through the MEG recordings and the MRI scans were analyzed to quantify the way in which the information is processed within the brain. For each frequency band, FC measures, named phase locking value (\({\text{PLV}}\)) [63] and magnitude coefficient (\({\text{MC}}\)) [64, 65], were computed starting from the combinations of pairs of signals derived from the 90 ROIs in which the brain cortex was parcellated. Details on the computation of individual FC measures are reported in Appendix.
Based on the time series used for the computation of the FC measures and on the averaging strategy along time, a set of seven different FC indices was obtained as follows.
Two representative sets of the band-filtered time series were considered: the cent signal and the pca signal. For the case of the cent signal, the geometrical centroid was computed for each ROI and the signal obtained from the closest source to the centroid was considered. To obtain the pca signal, the signals measured from the same brain area were subjected to a principal component analysis and the first principal component was considered. With the obtained combinations of signals, we extracted the FC measures for each pair of 4 s segment and finally obtained the average value along the segments.
An additional set of FC values was considered, the intra-ROI FC. In this case, the time series of all the sources pertaining to each ROI were used to estimate the FC indices among each combination of seed-test sources and finally a single average value has been extracted for each ROI.
Two different versions of the MC index, named \( {\text{MC}}_{{{\text{ma}}}} \) and \({\text{MC}}_{{{\text{am}}}}\), were derived with respect to the 4 s segments of the series. The \({\text{MC}}_{{{\text{ma}}}} \) was obtained by computing the mean of the Pearson complex correlation values among the segments first and then the absolute value, while the \({\text{MC}}_{{{\text{am}}}}\) was obtained by computing the absolute value of the complex Pearson correlation for each segment and then averaging the obtained results. The set of seven FC indices so derived are summarized in Table 2.
3.4 Derivation of image-based representations of FC
For each MEG sample and for a given a FC index, the measures computed between all possible pairs of ROIs, 90 in total, were topologically arranged into a 90 × 90 matrix. For each frequency band, seven FC maps corresponding to the seven FC indices, i.e., \({\text{PLV}}\_{\text{cent}}\), \({\text{PLV}}_{{{\text{pca}}}} \), \({\text{Intra}}\,{\text{ROI}} {\text{PLV}}\), \({\text{MC}}\_{\text{cent}}_{{{\text{ma}}}}\), \({\text{MC}}\_{\text{pca}}_{{{\text{ma}}}} ,{\text{MC}}\_{\text{cent}}_{{{\text{am}}}}\), and \({\text{MC}}\_{\text{pca}}_{{{\text{am}}}}\), with pixel values in the range [0,1] were derived. The θ, α, β, and β1 frequency bands were considered in this study, so that a total of 28 FC maps were generated per MEG sample. We rendered each map as a digital image, in which the topological arrangement of FC values and their spatial coordinates on the x-axis and the y-axis carry meaningful information. Such information, which is relative to the intricate communication patterns among neuronal populations, has not been fully investigated by previous MEG studies for AD diagnosis. In fact, MEG-based FC analysis has been addressed, up to now, by means of standard features-based approaches, without contemplating the spatial information contained in the FC maps [15,16,17, 36, 37]. In Fig. 2, three examples of \({\text{MC}}_{{\text{cent }}}\) maps in the β band are reported for a control case, as MCI patient and a pMCI patient, respectively. Although globally similar, it can be noted that the maps contain sub-regions of hyper or hypo-activation which provide information on both the amount of activation and the spatial location of the neuronal populations. Once derived for multiple frequency bands and for different FC indices, the set of images provides a direct visual representation of the neuronal activity ready to be decoded.
A further image-based representations, named RGB, was generated by combining multiple FC indices and frequency bands, as shown in the graphical representation of Fig. 3. In fact, given the symmetrical nature of the FC maps, data integration at the level of diagonal values and as triangular portions could reduce redundancy or lack of information. The discriminative power of different combination of features was checked at the classification level, and the best image-based representation was obtained by integrating the β1 sub-band frequency as the complementary triangular portion of the β band for the \({\text{MC}}\_{\text{cent}}\) indicator and by substituting the ones on the main diagonal in each color channel with the intra-ROI P \(\mathrm{LV}\) for the β1 sub-band frequency, as illustrated in Fig. 3. After distributing the two image-based representations of the \(\mathrm{MC}\_\mathrm{cent}\), i.e., \({\mathrm{MC}\_\mathrm{cent}}_{ma}\) and \({\mathrm{MC}\_\mathrm{cent}}_{\mathrm{am}}\), in different RGB channels, the RGB images so derived reduce redundancy by integrating multiple levels of data but all pertaining a similar information content in terms of frequency band. This choice, made at the pre-conversion stage, is consistent with the results reported by previous studies in the field [40]. In particular, β oscillations are believed to maintain the sensorimotor and cognitive state of an individual [41], with the motor performance that is impaired in AD, but not in MCI [42, 43], thus confirming the prominent role of the β band in the early detection of AD. We will see, in Sects. 4.1.1 and 4.1.2, that the phase synchrony in the β and β1 bands will be pivotal, also in the forms of individual FC maps, to discriminate the sMCI from the pMCI cases at the pre-conversion stage.
3.5 Deep-MEG feature transfer
The design of convolutional and pooling layers and their integration in deep learning architectures have boosted the performance of digital image classification in so many different scenarios, which have become the preferred choice for image analysis. In fact, with CNN, it is possible to extract meaningful image features automatically once the parameters of convolutional and pooling layers have been tuned and learned from big datasets of images, a procedure known as deep-feature transfer [5, 6]. Among the existing deep neural networks, AlexNet [49] is a large network structure with 60 million parameters and 650,000 neurons, consisting of five convolutional layers, most of them followed by max-pooling layers, and of three fully connected layers with a final 1000-way softmax layer for classification into 1000 classes. For our purposes, pre-trained AlexNet was used as a feature extractor without retraining the architecture, as shown in Fig. 1. After preliminary tests with other existing pre-trained CNNs providing comparable results, AlexNet was chosen due to its reduced number of intermediate descriptors. In particular, the pooled Conv5 layer was used to characterize the fine-grained structures present in MEG images and to decode, at the appropriate level of abstraction, the relatively simple patterns of interest [6].
For each patient, the image-based representations of FC were resized to a size of [227 × 227] pixels matrix using bicubic interpolation, and then, CNN feature transfer was performed using the pre-trained AlexNet architecture. The pooled Conv5 features so derived represent not only the individual values of the FC indicators for the relative frequency band but also their spatial arrangements and the generated patterns within the FC images. Dimensionality reduction on the features so derived was performed using standard deviation [66]. As the amount of dispersion of the deep features from their mean value should be indicative of higher information content and discrimination capability, only the features with a standard deviation higher than a given threshold were retained. The final subset of relevant features was selected using stepwise regression [68] with the training data of each round of cross-validation.
3.6 Classification
Linear discriminant analysis (LDA) and support vector machines (SVMs) were used as classification algorithms [43] with the scope of classifying MEG recordings relative to each patient as HC or sMCI or pMCI. For each frequency band and FC image, including the RGB images, the overall procedure was applied and the results obtained for each classification task are reported and discussed in Sects. 4 and 5 in terms of accuracy and area under the ROC curve. Leave-one-patient-out (LOPO) was used for cross-validation of results, and the classification was performed on a per-patient basis. Additional cooperative classification rules were designed to aggregate, at the test level, the assignment of base classifiers or ensemble modules trained with the image-based representations of FC. Further details are given in the following paragraph.
3.7 Cooperative classification
For the binary classification of MEG recordings as sMCI or pMCI, at the post- and pre-conversion stages, ensemble classifiers were derived to combine the probability scores of individual Deep-MEG modules (see ensemble #1 shown in Fig. 4a). For the more complex classification scenario including the MCI subjects at the pre-conversion stage and the HC subjects, a different ensemble architecture between two suboptimal binary classifiers was derived to aggregate the assessment of the individual Deep-MEG modules (ensemble #2 shown in Fig. 4b). With the second derived architecture, it was possible to detect the early signs of AD within a more complete scenario in which different rates of progression of the cognitive impairment (CI), going from absence of CI (in HC subjects) to pre-symptomatic AD phases (in pMCI subjects), were present.
Ensemble architectures proposed for classification of MEG recordings based on Deep-MEG features and image-based representations of FC. a Ensemble #1: ensemble architecture for binary classification. b Ensemble #2: cooperative architecture based on AND Logic and the RGB images for the three-class scenario of HC vs sMCI vs pMCI at the pre-conversion stage
3.7.1 Deep-MEG ensemble #1
An ensemble architecture was used in which base classifiers receive as input different FC images, also relative to diverse frequency bands, and are trained independently with the same set of patients. The outputs of individual classifiers, i.e., the probability scores of belonging to each class, are combined to derive the final assignment. In particular, the average values among the probability scores assigned to each class by the base classifiers were computed, and the sample was assigned to the class with the maximum obtained value between the two, as shown in Fig. 4a. Ensemble classifiers were obtained for discriminating the pMCI from the sMCI at both the pre- and post-conversion stages, as well as for discriminating HC from each of the two MCI classes.
3.7.2 Deep-MEG ensemble #2
The cooperative decision-making procedure, shown in Fig. 4b, is based on an AND logic between two base classifiers: one trained with the RGB images on MCI subjects for the discrimination of sMCI from pMCI patients and the other trained with the \({\text{PLV}}_{{{\text{cen}}}}\) map in the θ band, for the discrimination of HC from MCI patients. At the test level, a consensus mechanism is applied between the two classifiers so that the sample is assigned to the pMCI class only if both classifiers agree, i.e., if the probability scores of belonging to the pMCI and MCI classes are both higher than 0.5. The sample is assigned to the HC class if the probability score of belonging to the HC class is higher than 0.5; otherwise, the sample is assigned to the sMCI class.
The θ band has been chosen, in the present ensemble architecture, for the discrimination of HC from MCI patients due to its discrimination capability in preliminary tests and because changes in the θ band have been reported in the literature as indicative of MCI [39, 44]. In particular, the studies conducted by Lopez et al. [39, 44] outlined a hyper-synchronization of the θ band in MCI patients compared to the control subjects in resting state, which was also related to hippocampal atrophy and to lower global cognitive status. The increase in θ power is also considered as the most stable pattern of EEG activity in MCI patients [39], claim that has been confirmed by the present and the other studies on MEG signals [39, 44].
4 Results
In this section, the obtained results are presented for different classification scenarios. First, we report the results obtained for the classification of MEG recordings of the MCI subjects as sMCI or pMCI with respect to two classification approaches: (1) individual Deep-MEG classifiers based on different FC maps and on the RGB images (2) Deep-MEG ensemble #1. Finally, for the early detection of AD within a more complete scenario also including HC subjects, the results obtained with ensemble #2 are reported. Results are labeled as post-conversion when the MCI data include pMCI patients who met the criteria for probable AD and as pre-conversion when the pMCI patients were still clinically undistinguishable from the sMCI patients.
4.1 Classification of MEG recordings as sMCI or pMCI
4.1.1 Individual Deep-MEG modules
The results obtained with individual FC maps are first considered, and the frequency bands and image-based representations of FC that are relevant for each classification task, are reported.
In Fig. 5, we show the accuracy obtained at the post-conversion (a) and pre-conversion stages (b, c) for the classification of MEG recordings as sMCI or pMCI using LOPO. The results obtained with LDA and SVM and relative to the three best FC maps are reported for both scenarios in Fig. 5a, b. For the post-conversion stage, when the signs of AD are clinically evident in pMCI patients, accuracy values of 0.78, 0.87, and 0.70 were obtained with the θ, α, and β1 bands, respectively, and the \({\text{PLV}}\_{\text{pca}}\), \({\text{PLV}}\_{\text{cent}}\), and \({\text{MC}}\_{\text{cent}}_{{{\text{am}}}} \) FC maps, using LDA. The obtained values are reported together with the values of accuracy obtained using SVM (Fig. 5a). For the pre-conversion stage (Fig. 5b), accuracy values of 0.74, 0.78, and 0.76 were obtained with the β, β1, and β bands, respectively, and the \({\text{MC}}\_{\text{cent}}_{{{\text{ma}}}}\), \({\text{MC}}\_{\text{cent}}_{{{\text{am}}}} \), and \({\text{MC}}\_{\text{cent}}_{{{\text{am}}}}\) FC maps. For both post- and pre-conversion stages, the rest of FC maps or frequency bands provided lower results when decoded individually by a single classifier.
Results obtained with individual Deep-MEG modules. a, b Bar diagrams of the accuracy values obtained for classification of MEG recordings as sMCI or pMCI at the a post-conversion stage and b pre-conversion stage based on individual FC maps. For each stage, the results obtained with leave-one-patient-out cross-validation and relative to the three best FC maps are shown for LDA and SVM. c Confusion matrix obtained with the RGB images at the pre-conversion stage using a SVM classifier
For the pre-conversion stage, using a single classifier based on the RGB images as image representation of FC, accuracy values of 0.89 and 0.87 were obtained, respectively, with LDA and SVM. The confusion matrix obtained with LDA is reported in Fig. 5c. The FC indicators and relative frequency bands used to derive the RGB images are summarized in Table 3. The RGB-based Deep-MEG model was trained using three deep-features, on average, selected in each round of LOPO cross-validation.
4.1.2 Deep-MEG ensemble #1
Ensemble decisions were obtained by aggregating the probability scores of individual base classifiers (i.e., individual classifiers, LDA or SVM, of the Deep-MEG modules receiving as input a single FC map), as described in Sect. 3.6 and as illustrated in Fig. 4a. The FC maps performing the best with individual Deep-MEG modules in the post- and pre-conversion stages have been chosen to derive the corresponding ensembles. The FC indicators used to derive the image-based representations, or FC maps, are reported in Table 3: \({\text{MC}}\_{\text{cent}}_{{{\text{am}}}}\) in the β and β1 were used in the pre-conversion ensemble; \({\text{PLV}}\_{\text{cen}}\), \({\text{PLV}}\_{\text{pca}}\), and \( {\text{MC}}\_{\text{cent}}_{{{\text{am}}}}\), respectively, in the α, θ, and β1 frequency bands were selected for the post-conversion ensemble.
The contribution of the β and β1 bands at the pre-conversion stage confirms their role in the discrimination of sMCI from the pMCI cases [40,41,42,43]. Regarding the post-conversion stage, our analyses revealed that the phase synchrony in the α band serves as a predominant sign of AD only in symptomatic patients. In fact, a hyper-synchronization in the α band between the anterior cingulate region and the temporo-occipital region of pMCI patients as compared to sMCI was also reported by previous studies [16, 17] and seems to be correlated with cognitive performance. Two are the possible mechanisms behind such hyper-synchronization: (1) a compensation mechanism in response to the presence of compromised brain circuits in other brain areas; (2) the loss of GABAergic synapses caused by the accumulation of βamyloid plaques leading to establish aberrant relationships between the areas affected by the AD, which are hence the result of an inhibitory deficit [16]. The presence of the phase synchrony map in the θ band confirms, as reported in Sect. 3.7.2, its role in the recognition of MCI and, in this case, in the discrimination of sMCI from MCI progressed toward AD, i.e., pMCI at the post-conversion stage.
The results obtained for classification of MEG patients as sMCI or pMCI are shown in Fig. 6a in terms of accuracy for LDA and SVM base classifiers and in Fig. 6b and c in terms of confusion matrices. For the post-conversion stage, accuracy values of 0.93 and 0.85 were obtained with LDA and SVM, respectively (Fig. 6a). In Fig. 6b, the confusion matrix relative to LDA indicates a sensitivity of 0.93 for the pMCI cases, which correspond to patients with evident signs of AD, and specificity of 0.93. For the overall ensemble architecture, 42 deep features have been automatically selected, on average, to train the base classifiers.
Results obtained with the Deep-MEG ensemble #1. a Bar diagrams of the accuracy values obtained for classification of MEG recordings as sMCI or pMCI at the post-conversion stage (blue and orange) and at the pre-conversion stage. For each stage, the results obtained with LOPO cross-validation and relative to the three best FC maps are reported for LDA and SVM. b, c Confusion matrix at the post-conversion stage and pre-conversion stage obtained with Deep-MEG modules of LDA classifiers
For the pre-conversion stage, the histogram in Fig. 6a indicates the accuracy values of 0.89 and 0.87 obtained with the ensemble classification of MEG recordings as sMCI or pMCI with LDA and SVM, respectively. In this case, sensitivity of 0.89 and specificity of 0.78 were obtained, as reported in the confusion matrix in Fig. 6c, and 16 deep features were automatically selected, on average, to train the base classifiers.
4.2 Early prediction of AD
4.2.1 Deep-MEG ensemble #2
The aggregation method, named ensemble #2, was used for the classification scenario that included the HC subjects. An AND logic was used to derive the final assessment at the pre-conversion stage, as described in Sect. 3.7 and shown in Fig. 4b. In addition to the RGB images, which encode multiple FC indicators in the β and β1 bands, also the information of the \({\text{PLV}}_{{{\text{cent}}}}\) in the θ band was taken into account. The accuracy results obtained with ensemble #2 are reported in Fig. 6 relative to LDA.
For the three-class classification, an accuracy of 0.74 was obtained (Fig. 7a). For the overall ensemble, eight deep features were automatically selected, on average during the rounds of LOPO cross-validation, to train both base classifiers (three for the classifier receiving as input the RGB images and five for the classifier receiving as input the \({\text{PLV}}_{{{\text{cent}}}}\) maps in the θ band).
Results obtained with ensemble #2 for the early detection of AD. a Confusion matrix obtained for classification of MEG recordings as HC, sMCI, or pMCI. b ROC curves and corresponding AUC values obtained at pre-conversion stage using the two base classification modules composing the ensemble. c Confusion matrix obtained for classification of MEG recordings as pMCI or the rest. All the results are relative to the pre-conversion phase
The ROC curves relative to the individual base-ensemble classifiers, each trained to solve a specific binary classification task, are reported in Fig. 7b. An AUC of 0.90 was obtained for the classification of MEG recordings as sMCI or pMCI by the Deep-MEG module based on RGB images and an AUC of 0.83 was obtained for the classification of MEG recordings as HC or MCI using the \({\text{PLV}}_{{{\text{cent}}}}\) map in the θ band (Fig. 7b). Similar results were obtained using SVM. With the sole \( {\text{PLV}}_{{{\text{cent}}}}\) map in the θ band, when received as input by a single Deep-MEG-based classifier, accuracy value of 0.80 was obtained. For the discrimination of pMCI cases from the rest of the cases, i.e., HC + sMCI, accuracy of 0.87 and a sensitivity of detection of 0.82 were obtained with ensemble #2, as shown by the confusion matrix in Fig. 7c.
4.3 Comparative analysis
In this paragraph, the results obtained with the proposed approach are compared with the results obtained with the standard classification approach, i.e., when the FC indices were used as data for feature selection and classification without contemplating the information encoded in the spatial arrangement of pixel values. When the FC indicators were automatically selected at the training level using stepwise feature selection, we did not obtain any satisfactory results. To derive better results, the set of FC indicators with AUC values higher than a given threshold was considered to increase the classification performance of the standard approach and select the best combination of features. When single types of FC indicators were used, the higher results were 0.83 and 0.77 for the classification of sMCI and pMCI in the post- and pre-conversion phases, respectively, as compared with accuracy values of 0.87 and 0.78 obtained with individual Deep-MEG classifiers. In addition, for each classification scenario, multiple types of FC indicators and frequency bands were used and combined as a single feature vector. In this case, after selection based on AUC, accuracy values of 0.85 and 0.83 were obtained, for the classification of sMCI and pMCI in the post- and pre-conversion phases, respectively, as compared to accuracy values of 0.93 and 0.89 obtained with the proposed Deep-MEG approach. With the standard approach in the three-class scenario, we did not obtain satisfactory classification results. The best results obtained with the comparative analysis are reported in Table 4.
5 Discussion
We have presented a deep-feature transfer approach, named Deep-MEG, and a set of ensemble classification architectures for decoding MEG recordings based on a new visual perspective on FC for the early diagnosis of AD. Image-based representations of FC were derived starting from the MEG time series. The MEG signals were first processed and filtered to derive meaningful data for FC analysis and to quantify the spatiotemporal characteristics of the brain connectome, in conjunction with the anatomical information encoded in the MRI scans, as described in Sects. 3.1 and 3.2. Different versions of the \({\text{PLV}}\) and \({\text{MC}}\) indices were computed as FC descriptors and organized as RGB images, also relative to multiple frequency bands (see Sects. 3.3 and 3.4). Such images could be received as input data by the pre-trained CNN and pooling layers in the AlexNet network, used as feature extractors and decoders of FC patterns, as described in Sect. 3.5. Cooperative decision architectures among Deep-MEG modules allowed the integration of the brain signaling at multiple levels of frequency bands to derive increased performance (see Sect. 3.7).
The main novelty of the proposed study is the analysis of the MEG-FC patterns of the brain network via deep CNNs. We have shown that the information on the hypo- or hyper-synchronization is conveyed not only by the FC values, but it is also embedded in their spatial arrangement as FC maps, which gave us additional information on the connectome disruption related to AD. Individual Deep-MEG modules (see Sect. 4.1) allowed the discrimination of sMCI and pMCI patients at the post- and pre-conversion stages, with accuracy values of 0.87 and 0.78, respectively, using the \({\text{PLV}}\_{\text{cent}}\) in the α band and the \({\text{MC}}\_{\text{cent}}_{{{\text{am}}}} \) in the β1 band. For the binary classification of MEG recordings as HC or MCI, using a single Deep-MEG module based on the \({\text{PLV}}_{{{\text{cen}}}}\) map in the θ band, we obtained an accuracy of 0.80.
A composed image, named RGB, was designed to encode multiple levels of information also avoiding redundancies. The new set of deep features so extracted, boosted the classification performance at the pre-conversion stage to 0.89. In this scenario, we found that when data were integrated in different color channels of a single image, the encoded information has to be similar and homogeneous to guarantee appropriate decoding by the CNNs, i.e., indicators relative to the β and β1 bands.
As the connectivity patterns of different frequency bands were unique and differently informative in terms of activation patterns, multiband ensemble classifiers were used to integrate the information encoded in different image-based representations (see Sect. 4.2). By averaging the probability scores of the best image representations of FC at the decision level, increased accuracy values, i.e., 0.93 and 0.83 for the post- and pre-conversion stages, respectively, were obtained for the binary classification of MEG recordings as sMCI or pMCI. These results showed the fundamental role of integrating heterogeneous and diverse data (at the spatiotemporal and frequency levels) for better representation and decoding of the brain functional connectivity.
In our experiments in the three-class scenario also including the HC cases, none of the single image-based representations of FC was effective, neither was ensemble #1 among Deep-MEG modules based on LDA or SVM classifiers. More importantly, it was not possible to detect the pMCI cases at the pre-conversion stage, when also HC cases were present, i.e., the discrimination of pMCI cases from the HC and sMCI, that is the main goal for early detection of AD. The reasons of this finding may lie in the fact that the dynamic activity of the brain network in relation to diverse clinical conditions possesses diverse manifestations in terms of spatiotemporal and frequency responses and that the information relevant to each binary sub-problem are encoded into different frequency bands or FC indicators. Therefore, we used another ensemble logic, named ensemble #2, in which two classification modules solve the two different sub-tasks, as described in Sect. 3.6. Finally, we used an AND Logic to aggregate the assessment of the individual predictors. For the three-class results, accuracy value of 0.74 was obtained. The best discriminated class was the pMCI, with a percentage of detection of 0.82. The sMCI cases were mostly confounded with the HC cases. As the sMCI possess different severity levels, which may lie on a continuum from the cognitive perspective [68], different levels of cognitive impairments, in turn, may be associated with different FC connectivity patterns, some of which resulted to be similar to those of the HC cases. This is not surprising, especially considering the stability in terms of AD conversion of the sMCI cases over the three years of observation. When the HC and sMCI were considered as a single class (Fig. 7c), the accuracy of classification of ensemble #2 increased up to 0.87, maintaining a sensitivity for the pMCI class at 0.82. Ensemble #2 is the result of cooperation of only two Deep-MEG-based classifiers trained, on average, with eight deep features automatically selected during the rounds of LOPO cross-validation from the RGB images and the \({\text{PLV}}_{{{\text{cent}}}}\) map in the θ band. Data diversity and cooperation at the decision level were crucial to boost the recognition of pMCI cases at the pre-conversion stage and discriminate them from the HC and sMCI cases.
We have also shown that the proposed combination of deep spatiotemporal features and multiband ensemble classification showed superior performance as compared to other existing methods, including the standard approach to the analysis of FC, in which the FC indicators, also relative to different frequency bands, are used as feature descriptors. This was verified with respect to multiple combinations of FC features, even when the best FC features were selected on the complete pool based on their individual AUC values.
We tested multiple combinations of image representations of FC, and the higher results are reported in this study. A major advantage of this approach is that the learned models can be interpreted in neurophysiological terms. The results obtained with the present study support the notion of different functional brain connectivity patterns associated with different rates of progression and conversion to AD [68]. In line with other work in the literature [15, 16, 39,40,41,42,43,44], we have observed the role of specific frequency bands as potential biomarkers for the different phases of progression of the disease. In particular, a different set of FC images and frequency bands was determinant for the two ensemble classifiers relative to the post- and pre-conversion phases. In fact, at the post-conversion stage, our results indicate that the phase synchrony in the α band can serve as a predominant sign of AD in symptomatic patients [15, 16], while, at the pre-conversion stage, the results indicate evidence of changes in the pattern signs relative to the amplitude correlation in the β and β1 bands among the MEG signals [40,41,42,43]. Moreover, the discrimination of the HC cases from the MCI cases, instead, was favored by the presence of the FC indicators in the θ band [39, 44], which were not informative for the sMCI vs pMCI scenario in the pre-conversion phase (Table 3).
To further validate the platform, it would be important to test the proposed methods in a larger study. In the present work, to avoid overfitting, we extracted the deep data features from a pre-trained AlexNet architecture. Such features were automatically selected at the training level within rounds of LOPO cross-validation, thus allowing the training of simpler LDA or SVM classification modules based on a small set of features. In addition, our effort was devoted to identify the important MEG-based FC representations that inform classification (top–down approach) as the aggregation was performed using the best combinations of FC maps. The results obtained using knowledge-based computer vision techniques can be used as reference for deriving possible biomarkers for AD (down–up approach).
The results obtained in the present work compare favorably with the standard approach, in which the FC indicators are used as mono-dimensional training features, and with previous studies in the literature on the pre-conversion phase based on MEG data [17] or other imaging modalities [22, 23], posing the basis for further investigations on the proposed Deep-MEG architectures.
6 Conclusions
MEG provides the unique advantage of measuring the brain function with a remarkable combination of spatial and temporal resolutions. With this work, we have presented a novel system for decoding MEG recordings based on image-based representations of FC, deep CNN features, and ensemble classification architectures. The proposed methods for deriving and codifying the MEG-based FC measures allow the generation of pictures that represent, visually and numerically, the intricate communication patterns among spatially separated brain regions, which could be decoded by deep CNN features. The derivation of different cooperative architectures for integrating the spatiotemporal and multi-frequency information encoded in such images was the key to recognize the early alterations of the brain connectome relative to patients who undergo conversion to AD over a 3-year follow-up period.
In future, the analysis in resting state used in the present work may be extended with the analysis of other task-related activation patterns in order to optimize future applications of Deep-MEG architectures for predicting early signs of AD. Our findings may also have implications for the use of MEG-based FC as a biomarker in therapeutic trials. Finally, the proposed methods can be applied in other predictive scenarios to decode early signs of diverse neurodegenerative or neuropsychiatric diseases as well as to decode EEG signals.
References
Rajasree R, Columbus CC, Shilaja C (2020) Multiscale-based multimodal image classification of brain tumor using deep learning method. Neural Comput Appl. https://doi.org/10.1007/s00521-020-05332-5
Liu B, Liu Q, Zhu Z, Zhang T, Yang Y (2019) MSST-ResNet: deep multi-scale spatiotemporal features for robust visual object tracking. Knowl-Based Syst 164:235–252
Shi H, Qin C, Xiao D, Zhao L, Liu C (2020) Automated heartbeat classification based on deep neural network with multiple input layers. Knowl-Based Syst 188:105036
Jo T, Nho K, Saykin AJ (2019) Deep learning in Alzheimer’s disease: diagnostic classification and prognostic prediction using neuroimaging data. Front Aging Neurosci 11:220
Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Zheng L, Zhao Y, Wang S, Wang J, Tian Q (2016) Good practice in CNN feature transfer. arXiv preprint arXiv:1604.00133.
Polikar R (2012) Ensemble learning. Ensemble machine learning. Springer, Boston, MA, pp 1–34
Qi Z, Wang B, Tian Y, Zhang P (2016) When ensemble learning meets deep learning: a new deep support vector machine for classification. Knowl-Based Syst 107:54–60
Casti P, Mencattini A, Sammarco I, Velappa SJ, Magna G, Cricenti A, Di Natale C (2017) Robust classification of biological samples in atomic force microscopy images via multiple filtering cooperation. Knowl-Based Syst 133:221–233
Sengupta S, Basak S, Saikia P, Paul S, Tsalavoutis V, Atiah F, Peters A (2020) A review of deep learning with special emphasis on architectures, applications and recent trends. Knowl-Based Syst 194:105596
Craik A, He Y, Contreras-Vidal JL (2019) Deep learning for electroencephalogram (EEG) classification tasks a review. J Neural Eng 16(3):031001
Cheah KH, Nisar H, Yap VV, Lee CY (2020) Convolutional neural networks for classification of music-listening EEG: comparing 1D convolutional kernels with 2D kernels and cerebral laterality of musical influence. Neural Comput Appl 32(13):8867–8891
Oh SL, Hagiwara Y, Raghavendra U, Yuvaraj R, Arunkumar N, Murugappan M, Acharya UR (2018) A deep learning approach for Parkinson’s disease diagnosis from EEG signals. Neural Comput Appl 32(20):10927–10933
Yıldırım Ö, Baloglu UB, Acharya UR (2018) A deep convolutional neural network model for automated identification of abnormal EEG signals. Neural Comput Appl 32(20):15857–15868
López-Sanz D, Serrano N, Maestú F (2018) The role of magnetoencephalography in the early stages of Alzheimer’s disease. Front Neurosci 12:572
López ME, Bruna R, Aurtenetxe S, Pineda-Pardo JÁ, Marcos A, Arrazola J, Maestú F (2014) Alpha-band hypersynchronization in progressive mild cognitive impairment: a magnetoencephalography study. J Neurosci 34(44):14551–14559
Pusil S, López ME, Cuesta P, Bruña R, Pereda E, Maestú F (2019) Hypersynchronization in mild cognitive impairment: the ‘X’model. Brain 142(12):3936–3950
Boto E, Holmes N, Leggett J, Roberts G, Shah V, Meyer SS, Barnes GR (2018) Moving magnetoencephalography towards real-world applications with a wearable system. Nature 555(7698):657–661
Winblad B, Amouyel P, Andrieu S, Ballard C, Brayne C, Brodaty H, Fratiglioni L (2016) Defeating Alzheimer’s disease and other dementias: a priority for European science and society. Lancet Neurol 15(5):455–532
Petersen RC (2004) Mild cognitive impairment as a diagnostic entity. J Intern Med 256(3):183–194
Palmqvist S, Mattsson N, Hansson O, Alzheimer’s Disease Neuroimaging Initiative (2016) Cerebrospinal fluid analysis detects cerebral amyloid-β accumulation earlier than positron emission tomography. Brain 139(4):1226–1236
Suk HI, Lee SW, Shen D, Initiative ADN (2014) Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage 101:569–582
Lu D, Popuri K, Ding GW, Balachandar R, Beg MF (2018) Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural MR and FDG-PET images. Sci Rep 8(1):1–13
Choi H, Jin KH, Alzheimer’s Disease Neuroimaging Initiative (2018) Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav Brain Res 344:103–109
Fan Z, Xu F, Qi X, Li C, Yao L (2019) Classification of Alzheimer’s disease based on brain MRI and machine learning.Neural Comput Appl 32(20):1927–1936
Gaubert S, Raimondo F, Houot M, Corsi MC, Naccache L, Diego Sitt J, Dubois B (2019) EEG evidence of compensatory mechanisms in preclinical Alzheimer’s disease. Brain 142(7):2096–2112
Hojjati SH, Ebrahimzadeh A, Khazaee A, Babajani-Feremi A, Alzheimer’s Disease Neuroimaging Initiative (2017) Predicting conversion from MCI to AD using resting-state fMRI, graph theoretical approach and SVM. J Neurosci Methods 282:69–80
Li R, Rui G, Zhao C, Wang C, Fang F, Zhang Y (2019) Functional network alterations in patients with amnestic mild cognitive impairment characterized using functional near-infrared spectroscopy. IEEE Transactions on Neural Systems and Rehabilitation Engineering.
Croce P, Zappasodi F, Marzetti L, Merla A, Pizzella V, Chiarelli AM (2018) Deep Convolutional Neural Networks for feature-less automatic classification of Independent Components in multi-channel electrophysiological brain recordings. IEEE Trans Biomed Eng 66(8):2372–2380
Zubarev I, Zetter R, Halme HL, Parkkonen L (2019) Adaptive neural network classifier for decoding MEG signals. Neuroimage 197:425–434
Aoe J, Fukuma R, Yanagisawa T, Harada T, Tanaka M, Kobayashi M, Inoue Y, Yamamoto S, Ohnishi Y, Kishima H (2019) Automatic diagnosis of neurological diseases using MEG signals with a deep neural network. Sci Rep 9(1):1–9
Tokozume Y, Harada T (2017) Learning environmental sounds with end-to-end convolutional neural network. In: 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE pp. 2721–2725
Amezquita-Sanchez JP, Adeli A, Adeli H (2016) A new methodology for automated diagnosis of mild cognitive impairment (MCI) using magnetoencephalography (MEG). Behav Brain Res 305:174–180
Lopez-Martin M, Nevado A, Carro B (2020) Detection of early stages of Alzheimer’s disease based on MEG activity with a randomized convolutional neural network. Artif Intell Med 107:101924
Filippi M, Agosta F, Scola E, Canu E, Magnani G, Marcone A, Valsasina P, Caso F, Copetti M, Comi G, Cappa SF, Falini A (2013) Functional network connectivity in the behavioral variant of frontotemporal dementia. Cortex 49(9):2389–2401
Gómez C, Juan-Cruz C, Poza J, Ruiz-Gómez SJ, Gomez-Pilar J, Núñez P, García M, Fernández A, Hornero R (2018) Alterations of effective connectivity patterns in mild cognitive impairment: an meg study. J Alzheimers Dis 65(3):843–854
Yu M, Engels MMA, Hillebrand A, van Straaten ECW, Gouw AA, Teunissen C, van der Flier WM, Scheltens P, Stam CJ (2017) Selective impairment of hippocampus and posterior hub areas in Alzheimer’s disease: an MEG-based multiplex network study. Brain 140:1466–1485
Pusil S, Dimitriadis SI, López ME, Pereda E, Maestú F (2019) Aberrant MEG multi-frequency phase temporal synchronization predicts conversion from mild cognitive impairment-to-Alzheimer’s disease. NeuroImage Clin 24:101972
López ME, Cuesta P, Garcés P, Castellanos PN, Aurtenetxe S, Bajo R, Marcos A, Delgado ML, Montejo P, López-Pantoja JL, Maestú F, Fernandez A (2014) MEG spectral analysis in subtypes of mild cognitive impairment. Age 36(3):9624
Poil SS, De Haan W, van der Flier WM, Mansvelder HD, Scheltens P, Linkenkaer-Hansen K (2013) Integrative EEG biomarkers predict progression to Alzheimer’s disease at the MCI stage. Front Aging Neurosci 5:58
Engel AK, Fries P (2010) Beta-band oscillations—signalling the status quo? Curr Opin Neurobiol 20(2):156–165
Sheridan PL, Solomont J, Kowall N, Hausdorff JM (2003) Influence of executive function on locomotor function: divided attention increases gait variability in Alzheimer’s disease. J Am Geriatr Soc 51(11):1633–1637
Pettersson AF, Olsson E, Wahlund LO (2005) Motor function in subjects with mild cognitive impairment and early Alzheimer’s disease. Dement Geriatr Cogn Disord 19(5–6):299–304
López ME, Garcés P, Cuesta P, Castellanos NP, Aurtenetxe S, Bajo R, Sancho M (2014) Synchronization during an internally directed cognitive state in healthy aging and mild cognitive impairment: a MEG study. Age 36(3):9643
Montine TJ, Monsell SE, Beach TG, Bigio EH, Bu Y, Cairns NJ, Lee EB (2016) Multisite assessment of NIA-AA guidelines for the neuropathologic evaluation of Alzheimer’s disease. Alzheimers Dement 12(2):164–169
Dubois B, Albert ML (2004) Amnestic MCI or prodromal Alzheimer’s disease? Lancet Neurol 3(4):246–248
Casti P, Giovannetti A, Susi G, Mencattini A, Pusil SA, García MEL, Di Natale C, Martinelli E (2020) A deep CNN-based approach for predicting MCI to AD conversion: developing topics. Alzheimer’s Dement 16:e047570
Sporns O, Tononi G, Kötter R (2005) The human connectome: a structural description of the human brain. PLoS Comput Biol 1(4):e42
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25:1097–1105
Bishop CM (2006) Pattern recognition and machine learning. Springer
Oldfield RC (1971) The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia 9(1):97–113
Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Snyder PJ (2011) The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7(3):270–279
McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr, Kawas CH, Mohs RC (2011) The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7(3):263–269
Hachinski VC, Iliff LD, Zilhka E, Du Boulay GH, McAllister VL, Marshall J, Symon L (1975) Cerebral blood flow in dementia. Arch Neurol 32(9):632–637
Bai Y, Hu Y, Wu Y, Zhu Y, He Q, Jiang C, Fan W (2012) A prospective, randomized, single-blinded trial on the effect of early rehabilitation on daily activities and motor function of patients with hemorrhagic stroke. J Clin Neurosci 19(10):1376–1379
Oostenveld, R., Fries, P., and Maris, E. (2011). Schoffelen JrM (2010) FieldTrip: Open Source Software for Advanced Analysis of MEG. EEG, and Invasive Electrophysiological Data. computational Intelligence and Neuroscience.
Robinson SE (2004) Localization of event-related activity by SAM (erf). Neurol Clin Neurophysiol NCN 2004:109–109
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Joliot M (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15(1):273–289
Taulu S, Simola J (2006) Spatiotemporal signal space separation method for rejecting nearby interference in MEG measurements. Phys Med Biol 51(7):1759
Fischl B (2012) FreeSurfer. Neuroimage 62(2):774–781
Nolte G (2003) The magnetic lead field theorem in the quasi-static approximation and its use for magnetoencephalography forward calculation in realistic volume conductors. Phys Med Biol 48(22):3637
Van Veen BD, Van Drongelen W, Yuchtman M, Suzuki A (1997) Localization of brain electrical activity via linearly constrained minimum variance spatial filtering. IEEE Trans Biomed Eng 44(9):867–880
Lachaux JP, Rodriguez E, Martinerie J, Varela FJ (1999) Measuring phase synchrony in brain signals. Hum Brain Mapp 8(4):194–208
Bierstedt SE, Hünicke B, Zorita E (2015) Variability of wind direction statistics of mean and extreme wind events over the Baltic Sea region. Tellus A Dyn Meteorol Oceanogr 67(1):29073
Von Storch H, Zwiers FW (2001) Statistical analysis in climate research. Cambridge University Press
Yousefpour A, Ibrahim R, Hamed HNA, Hajmohammadi MS (2014) Feature reduction using standard deviation with different subsets selection in sentiment analysis. Asian Conference on Intelligent Information and Database Systems. Springer, Cham, pp 33–41
Draper NR, Smith H (1998) Applied regression analysis, vol 326. Wiley, Hoboken
Petrella JR, Sheldon FC, Prince SE, Calhoun VD, Doraiswamy PM (2011) Default mode network connectivity in stable vs progressive mild cognitive impairment. Neurology 76(6):511–517
Gabor D (1946) Theory of communication Part 1: the analysis of information. J Inst Electr Eng Part III Radio Commun Eng 93(26):429–441
Stam CJ, Nolte G, Daffertshofer A (2007) Phase lag index: assessment of functional connectivity from multi channel EEG and MEG with diminished bias from common sources. Hum Brain Mapp 28(11):1178–1193
Acknowledgements
This study was supported by two projects from the Spanish Ministry of Economy and Competitiveness (PSI2009-14415-C03-01 and PSI2012-38375-C03-01).
Funding
Open access funding provided by Università degli Studi di Roma Tor Vergata within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
1.1 Derivation of the FC indices
We initially computed the analytic signal associated to the time series based on the Hilbert transform.
The analytic signal, \(s_{AN} \left( t \right)\), of a real-valued signal, \( s\left( t \right)\), is a complex-valued signal defined as [69, 70]:
where the real part corresponds to the original signal, and the imaginary part, \(s_{H} \left( t \right)\), is the Hilbert transform of \(s\left( t \right)\), defined as:
with PV the Cauchy principal value. From Eq. (A1), \(A_{s} \left( t \right)\) and \(\varphi_{s} \left( t \right){ }\) are the instantaneous amplitude and the instantaneous phase, respectively, of the analytic signal.
The Phase Locking Value (\({\text{PLV}}\)) [63] was computed as FC measure of phase synchrony, since it quantifies how the phase difference between two signals is preserved during the time course. First, the instantaneous phase \(\varphi \left( t \right)\) has been extracted for each of the 4 s segment of the series, s(t). Finally, starting from pairs of analytic signals, The PLV between two time series was evaluated with the following expression:
where \(N\) indicates the number of time points, 〈∙〉 the time average and \(\Delta \varphi_{{{\text{rel}}}} \left( {t_{n} } \right)\) the cyclic relative phase, i.e., the difference between the instantaneous phases of the two signals, bounded in the interval [0-2π). \({\text{PLV}}\) ranges from 0 to 1; a value close to 0 reflects the relative phase is uniformly distributed (or the phase distribution has n peaks at values which differ by (2π)/n), while a value of 1 indicates perfect phase locking between the time series.
As a second index of co-variability between two signals, we evaluated the magnitude of the complex Pearson correlation between the analytic signals associated to the time series [64, 65], that we refer to as the Magnitude Coefficient (\({\text{MC}}\)):
where the superscript * denotes the complex conjugate. The MC gives a measure of the strength of the linear relationship between the envelopes of the signals, in a scale that ranges from 0 to 1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Giovannetti, A., Susi, G., Casti, P. et al. Deep-MEG: spatiotemporal CNN features and multiband ensemble classification for predicting the early signs of Alzheimer’s disease with magnetoencephalography. Neural Comput & Applic 33, 14651–14667 (2021). https://doi.org/10.1007/s00521-021-06105-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-06105-4