Abstract
Purpose: Lobular breast cancer (LBC) accounts for ~ 15% of breast cancer. Here, we studied the frequency of pathogenic germline variants (PGVs) in an extended panel of genes in women affected with LBC. Methods: 302 women with LBC and 1567 without breast cancer were tested for BRCA1/2 PGVs. A subset of 134 LBC affected women who tested negative for BRCA1/2 PGVs underwent extended screening, including: ATM, CDH1, CHEK2, NBN, PALB2, PTEN, RAD50, RAD51D, and TP53. Results: 35 PGVs were identified in the group with LBC, of which 22 were in BRCA1/2. Ten actionable PGVs were identified in additional genes (ATM(4), CDH1(1), CHEK2(1), PALB2(2) and TP53(2)). Overall, PGVs in three genes conferred a significant increased risk for LBC. Odds ratios (ORs) were: BRCA1: OR = 13.17 (95%CI 2.83–66.38; P = 0.0017), BRCA2: OR = 10.33 (95%CI 4.58–23.95; P < 0.0001); and ATM: OR = 8.01 (95%CI 2.52–29.92; P = 0.0053). We did not detect an increased risk of LBC for PALB2, CDH1 or CHEK2. Conclusion: The overall PGV detection rate was 11.59%, with similar rates of BRCA1/2 (7.28%) PGVs as for other actionable PGVs (7.46%), indicating a benefit for extended panel genetic testing in LBC. We also report a previously unrecognised association of pathogenic variants in ATM with LBC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Invasive lobular cancer (ILC) is the most common special histological subtype of breast cancer representing 10–15% of cases overall. ILC is associated with a higher risk of inheritance than invasive ductal cancers (IDC) which are also termed 'no special type' and represent 70% of cases overall [1, 2]. Pathogenic germline variants (PGVs) in one of the known high-risk breast cancer susceptibility genes e.g. BRCA1, BRCA2, PALB2 confer a high risk of developing breast cancer and are enriched in familial cases. Notably, lobular breast cancer (LBC) is less common as a proportion of breast cancers in women with BRCA1 PGVs compared to women without BRCA1 PGVs [3,4,5,6] As the main pathway to breast cancer is from basal progenitor cells leading to the predominant ductal triple negative breast cancer, this route excludes the predominantly estrogen receptor positive lobular cancer. However, individuals with BRCA2 PGVs have a similar proportion of LBC to women without BRCA2 PGVs. Familial breast cancer not associated with variants in BRCA1 or BRCA2 is more likely to be lobular than those with a BRCA1 or BRCA2 variant [7, 8]. In our first pathology update of the Manchester Scoring system, 11% of familial breast cancers that tested negative for BRCA1/2 were lobular, but lobular cancer was present in only 1.6% of index BRCA1 cases [8].
A recent increase in the use of multi-gene panels to screen for breast cancer-associated gene variants beyond BRCA1 and BRCA2 has started to identify PGVs in additional genes. Several studies have reported the association for the diffuse gastric cancer predisposition gene, CDH1, in lobular, rather than ductal breast cancer [9,10,11,12]. Petridis et al. screened six genes for an association with LBC and found that variants in BRCA2, CHEK2, and PALB2 in addition to CDH1 were all enriched in women with lobular compared to ductal cancer [5].
Only PGVs in the CDH1 gene have been convincingly shown to cause a specific high risk of LBC with lifetime risks of about 40% in women [13, 14]. However, CDH1 germline PGVs are very rare with a population incidence of 1 in 10–100,000 [15]. Indeed, a study of 1434 women with lobular breast cancer only found five (0.35%) with a CDH1 PGV [5]. As such there are still many individuals with LBC and a family history of breast cancer in whom no PGV has been identified.
Here, we present an analysis of the frequency of detectable pathogenic variants including those in an extended panel of nine non-BRCA1/2 genes in a cohort of 302 women with LBC.
Methods
Patient materials
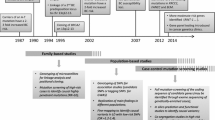
Women were eligible for this study if they had a histologically confirmed diagnosis of LBC (lobular invasive lobular carcinoma). A total of 302 women affected with LBC were included in the study. Two-hundred and sixty four of these women were seen at the Manchester Centre for Genomic Medicine (MCGM) and 259 (98.1%) of these had a family history of breast or ovarian cancer or a second primary breast/ovarian cancer. The first families ascertained through MCGM were from 1990, although testing for BRCA1/2 only started in 1996. A further 38 women took part in the population-based study, Predicting the Risk Of Cancer At Screening (PROCAS), held in Greater Manchester [16]. Additionally, 1567 women without a breast cancer diagnosis at entry (aged 46–73 years) who were also recruited to the PROCAS study were included as controls. To obtain a population average dataset we included 124 women who subsequently developed breast cancer to provide an ~ 8% population risk of breast cancer at a median last age of follow up of 69 years. Clinical or research consent was given for extended testing of breast cancer associated genes (approval from the North Manchester Research Ethics Committee (reference 09/H1008/81(PROCAS) and 08/H1006/77)).
Genetic screening
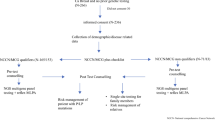
For women that were seen at the MCGM, DNA was extracted from lymphocytes and all these samples were initially sequenced for PGVs in BRCA1/2 by a combination of Next Generation Sequencing and MLPA. A subset of 134 of the patients that did not harbour a PGV in either of those genes underwent extended testing. All women recruited through PROCAS provided a saliva sample for DNA extraction. They underwent panel testing as part of the Breast Cancer Risk after Diagnostic Gene Sequencing (BRIDGES) study [17]. The BRIDGES study performed sequencing of 33 genes (AKT1, ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, FAM175A, FANCC, FANCM, GEN1, MEN1, MRE11A, MSH2, MSH6, MUTYH, NBN, NF1, PALB2, PIK3CA, PMS2, PPM1D, PTEN, RAD50, RAD51C, RAD51D, RECQL, RINT1, STK11, TP53 and XRCC2). Forty-six women underwent clinical panel testing (the panel included: ATM, BAP1, BRCA1, BRCA2, CDH1, CHEK2, MLH1, NBN, PALB2, PTEN, RAD50, RAD51D, RECQL, TP53, and for 20 of these women BARD1, MSH6, MSH2 and PMS2 were also included) and 50 women underwent targeted exome sequencing through the Beijing Genomics Institute (BGI), Shenzhen, Guangdong, China. A minimum of nine genes were screened in each patient that underwent extended testing. These genes were: ATM, CDH1, CHEK2, NBN, PALB2, PTEN, RAD50, RAD51D, and TP53 (Supplemental Fig. 1).
Samples that underwent panel testing though BRIDGES did not undergo copy number analysis. The samples that underwent targeted exome sequencing only had copy number testing for BRCA1/2.
Variants were classified according to the ACMG guidelines [18]. All identified pathogenic variants were confirmed through the clinical diagnostics laboratory. Only variants that were classified as ‘likely pathogenic’ or ‘pathogenic’ are reported here.
Tumour pathology information was obtained for each case through hospital records, and cancer registries as previously described [19]. The probability of a BRCA1/2 PGV was determined using the Manchester score (MS) for each affected individual [20]. Genes were considered actionable for breast cancer if they had published data confirming at least a twofold relative risk of breast cancer [21]. Associations between LBC and PGVs in the screened genes were calculated by Fisher’s exact test using GraphPad Prism 8.
Results
Pathogenic variants
A total of 302 women affected with invasive LBC were initially screened for PGVs in BRCA1/2. Within this group 22 (7.28%) PGVs were identified in BRCA1/2 (5 in BRCA1 (1.66%) and 17 in BRCA2 (5.63%)). Of the 280 women who tested negative for BRCA1/2 PGVs, 134 (47.9%) had sufficient DNA available for extended panel testing of at least ATM, CDH1, CHEK2, NBN, PALB2, PTEN, RAD50, RAD51D, and TP53. This resulted in the detection of an additional 13 PGVs of which 10 (7.46%) could be considered to be actionable with at least a confirmed two fold increased breast cancer risk [21] (4 in ATM, 1 in CDH1, 1 in CHEK2, 2 in PALB2, and 2 in TP53) (Table 1). Additional PGVs genes with a less clear association with breast cancer were found in MSH6, RAD50, and NBN. Thus, in total, PGVs were detected in 35 individuals (11.59%). MSH6 was not assayed on every clinical gene panel and was therefore screened in 108 of the 134 cases. Twenty-five women with lobular carcinoma in situ were also screened for PGVs, but none of them carried a PGV.
In the control group of 1567 women, a total of 36 PGVs (2.30%) were detected. A first degree family history of breast cancer was present in 11.9% of PROCAS controls who provided a saliva DNA (and were analysed in BRIDGES) compared to 11.3% of PROCAS controls who did not provide a DNA sample. This indicates that there is no bias towards family history in providing DNA samples. There were eleven PGVs in BRCA1/2 (2 in BRCA1 and 9 in BRCA2), as well as six in ATM, six in CHEK2, three in PALB2, three in NBN, one in MSH6, and six in RAD50.
Odds ratios (ORs) for each gene are presented in Table 2. The ORs for BRCA2 (OR = 10.33 (95%CI 4.58–23.95; P < 0.0001)), BRCA1 (OR = 13.17 (95%CI 2.83–66.38; P = 0.0017)), and ATM (OR = 8.01 95%CI 2.52–29.92; P = 0.0053) confirmed elevated LBC risks with the lower 95% confidence intervals (CI) above twofold. We were unable to confirm this increase in LBC risk for PALB2 and CHEK2. CDH1 was non assessable due to absence of a reliable population incidence and the identification of only one case, previously reported [22]. All patients were also tested for CHEK2 c.1100delC. We only identified one woman with LBC and six controls with this PGV (OR = 0.86; 95%CI 0.08–5.23; P ≥ 0.9999).
ATM variant segregation
One of the individuals with an ATM PGV did not have a family history of breast cancer. For only one family, an additional family member was available to perform segregation analysis. This family member was affected with ductal breast cancer rather than lobular breast cancer and also carried the same ATM PGV. The other two families also had mixed lobular and ductal pathologies.
Receptor status
Of the 302 women, receptor status was known for 220 (72.82%). The majority were estrogen receptor (ER) positive (214 cases, 97.27%). Of the ER positive cases, HER2 receptor status was also known in 137 cases and, of these, 128 (93.43%) were HER2-negative (Table 3).
In three of the only six ER negative cases, a PGV was present (BRCA1(1); BRCA2(1); MSH6(1)). The majority of PGVs were identified in women with ER positive (14 PGVs: BRCA1(2); BRCA2(7); TP53(1); ATM(2); CDH1(1); CHEK2(1)) or ER positive/HER2- negative receptor status (11 PGVs: BRCA1(1); BRCA2(6); PALB2(1); ATM(2); RAD50(1)). One PGV was found in a woman with ER positive/HER2 positive tumour receptor status (BRCA2) and six PGVs (BRCA1(1); BRCA2(2); TP53(1); PALB2(1); NBN(1)) were found in women whose receptor status was unknown (Table 3).
Manchester score
In order to assess the probability of PGVs in BRCA1/2, MS was determined for all affected women. The majority of women had a MS < 20 (232/302) and a detection rate of 4.74%. The detection rate of BRCA1/2 PGVs in women with a MS > 20 was 15.71% (Table 3).
Only 43 of the 302 (14.23%) women did not have a family history of breast cancer and four of these carried an actionable PGV (1 in BRCA1, 1 in BRCA2 and 1 in TP53 and 1 in ATM). The BRCA1 and BRCA2 PGVs were identified in women diagnosed with a first primary lobular cancer who then developed a second primary breast or ovarian cancer, respectively, whereas the TP53 PGV was identified in a woman with a first primary lobular breast cancer at age 21 who then developed bilateral disease.
Discussion
In this cohort of 302 women diagnosed with LBC, we identified a PGV in 11.59%. Interestingly, the detection rate of PGVs beyond BRCA1/2 was 7.46%, which is similar to the BRCA1/2 detection rate (7.28%). Although the overall rate was low, the comparable rates of detection between BRCA1/2 and non-BRCA1/2 variants implies that extended testing may be particularly beneficial in women with LBC and a family history of breast cancer.
We identified four PGVs in ATM, which is equal to the number of PGVs detected in BRCA1 even though ATM was screened in less than half the individuals where BRCA1 was tested. The OR of 8.01 and two fold increased risk at the lower end of the confidence interval supports an association of ATM PGVs with LBC, which is consistent with Lu et al. [23] This group identified six PGVs in ATM in 369 (1.63%) patients with LBC (OR = 3.50; 95%CI 1.10–9.73), although this is a lower frequency than in our study (4/134 (2.99%)).
In line with previous observations, the four ATM PGVs identified in this study were found in women with ER + (/HER-) tumour characteristics [24, 25], which is also the most common tumour type seen in LBC [26].
In this study, we found as many PGVs in ATM as in PALB2 and CHEK2 combined. The recent large study by Petridis et al. [5] found that BRCA2, PALB2 and CHEK2 PGVs were the most prevalent in women affected with LBC [5]. However, ATM was not investigated in the Petridis study. A recent large American-based study of germline genetic testing criteria in 3907 women with breast cancer, identified 43 ATM variants, but did not distinguish between women with ductal and lobular cancers [27].
CDH1 is known to predispose to diffuse gastric cancer and has more recently been associated with LBC [5, 12]. In our cohort, we only identified one CDH1 PGV in a LBC family and there was no history of gastric cancer in this family. Another recent study reported an association with MSH6 (PGV identified in 7 of 590 patients) [23], although this was questioned as being due to potential sequencing errors [28]. We were not able to evaluate this fully as MSH6 variants were screened in only 80.6% of the samples tested and our study was not powered to refute this association.
We identified two cases with a TP53 PGV. This is in contrast to Petridis et al. who did not identify any TP53 PGVs in an unselected series of 1434 lobular cancers [5]. A lack of association with lobular cancer and germline TP53 was also suggested by Ditchi et al. in 2019 [6] but this is based on one case in only 57 carriers and the 95% CI do not exclude a 10% rate. One of our women was diagnosed with very early onset, bilateral breast cancer (the contralateral tumour was reported as invasive ductal carcinoma histologically). Very early onset ductal breast cancer is associated with TP53 PGVs and therefore potentially explains the TP53 PGV in this lobular case [29]. Given the extremely high odds ratios for TP53 in very early onset bilateral and familial cases an increased risk of lobular cancer is nonetheless still possible.
Interestingly, an MS of ≥ 20 is usually associated with a probability of BRCA1/2 involvement of around 35%; we only identified BRCA1/2 PGVs in 11/75 (14.67%) in women with a MS ≥ 20 despite the MS already including a downward adjustment for LBC [8]. This almost certainly indicates that even the current reduction in score for lobular breast cancer of -2 is insufficient to reflect BRCA1 risk. The absence of an association of LBC with BRCA1 has already been noted by ourselves and others [6], with only 2/342 (0.58%) LBCs in BRCA1 breast cancer patients in one study [6]. The higher odds ratio for BRCA1 in our study is therefore likely to be explained by the strong family history of breast and/or ovarian cancer in relatives rather than a true association with BRCA1. Although the MS is specifically used for BRCA1/2, it does contain an element of assessing high risk for inherited breast cancer by scoring all breast cancers in the lineage and increasing the score in younger cases. Inclusion of the remaining genes only increased this to 20% with an identified PGV. The relative absence of family histories of ovarian cancers in the series (only 43/302 and 8/134 of those with panel testing) would suggest that the absence of PGVs in so many with high MS is likely due to missing heritability. This indicates there is still a greater proportion of the heritability unexplained for lobular cancer.
Although we have confirmed associations with well described breast cancer genes and lobular cancer, most of the ORs in our study and the literature are in keeping with the ORs for overall breast cancer risk, which does not suggest that these genes have a specific link to lobular cancers. In particular the OR of only 0.86 for c.1100delC in CHEK2 is less than would be expected for ER + ductal cancer and does not support CHEK2 being particularly associated with LBC. However, the ORs for ATM of 8.01 fold in this study and 3.50 fold from a previous study [23] if confirmed in further work would imply a more specific link to lobular cancers. The ORs for BRCA1 in contrast were as high as expected for overall breast cancer risk; however this may reflect the rather low carrier frequency in the control population of only two of 1567 individuals, as well as the strong family histories of breast and/or ovarian cancer.
One of the limitations of this study was that we were unable to account for any copy number variations (CNVs) in samples tested through the BRIDGES study. Therefore, the results for some of the genes may be underestimated. For example, we have estimated that CNVs in BRCA1 account for 20% of the PGVs in non-Jewish families [30]. We also did not have detailed pathology of the breast cancers in the families of those with familial breast cancer, although a number of families did have confirmed lobular family history, including the family with the CDH1 variant. Also, as only a subset of the full cohort has been tested for the extended gene panel, there may be still undetected PGVs. In contrast to many other studies, we have been able to use local control samples rather than infer odds ratios from frequencies on gnomAD [31]. The relatively high frequency of BRCA1/2 in PROCAS controls of 0.56% does not appear to be due to bias as 1st degree breast cancer family history was not more frequent in subjects who provided a DNA sample. It is also very similar to a study of 50,726 adult biobank volunteers in the USA which found a 0.5% rate [32]. Another possible limitation is that there was no central pathology review. However, both of our TP53-positive cases were reviewed as part of their treatment at a major cancer centre (the Christie) and our study reflects actual practice, as pathologies are not usually reviewed in order to undertake genetic testing.
In summary, we have identified an association between ATM and an increased risk of LBC, but it is likely that further familial lobular cancer genes remain to be discovered as only 20% of patients with LBC and a family history of breast cancer with MS ≥ 20 were explained by currently known breast cancer predisposition genes.
Data availability
The datasets analysed during the current study are available from the corresponding author on reasonable request.
References
Rosen PP, Lesser ML, Senie RT, Kinne DW (1982) Epidemiology of breast carcinoma III: relationship of family history to tumor type. Cancer 50(1):171–179. https://doi.org/10.1002/1097-0142(19820701)50:1%3c171::aid-cncr2820500132%3e3.0.co;2-m
Cannon-Albright LA, Thomas A, Goldgar DE, Gholami K, Rowe K, Jacobsen M, McWhorter WP, Skolnick MH (1994) Familiality of cancer in Utah. Cancer Res 54(9):2378–2385
Fountzilas E, Konstantopoulou I, Vagena A, Apostolou P, Papadimitriou C, Christodoulou C, Tryfonopoulos D, Manousou K, Delimitsou A, Papamentzelopoulou M, Fountzilas G, Yannoukakos D, Fostira F (2019) Pathology of BRCA1- and BRCA2-associated breast cancers: known and less known connections. Clin Breast Cancer. https://doi.org/10.1016/j.clbc.2019.08.003
Lakhani SR, Gusterson BA, Jacquemier J, Sloane JP, Anderson TJ, van de Vijver MJ, Venter D, Freeman A, Antoniou A, McGuffog L, Smyth E, Steel CM, Haites N, Scott RJ, Goldgar D, Neuhausen S, Daly PA, Ormiston W, McManus R, Scherneck S, Ponder BA, Futreal PA, Peto J, Stoppa-Lyonnet D, Bignon YJ, Stratton MR (2000) The pathology of familial breast cancer: histological features of cancers in families not attributable to mutations in BRCA1 or BRCA2. Clin Cancer Res 6(3):782–789
Petridis C, Arora I, Shah V, Moss CL, Mera A, Clifford A, Gillett C, Pinder SE, Tomlinson I, Roylance R, Simpson MA, Sawyer EJ (2019) Frequency of Pathogenic Germline Variants in CDH1, BRCA2, CHEK2, PALB2, BRCA1, and TP53 in Sporadic Lobular Breast Cancer. Cancer Epidemiol Biomarkers 28(7):1162–1168. https://doi.org/10.1158/1055-9965.EPI-18-1102
Ditchi Y, Broudin C, El Dakdouki Y, Muller M, Lavaud P, Caron O, Lejri D, Baynes C, Mathieu MC, Salleron J, Benusiglio PR (2019) Low risk of invasive lobular carcinoma of the breast in carriers of BRCA1 (hereditary breast and ovarian cancer) and TP53 (Li-Fraumeni syndrome) germline mutations. Breast J 25(1):16–19. https://doi.org/10.1111/tbj.13154
Dossus L, Benusiglio PR (2015) Lobular breast cancer: incidence and genetic and non-genetic risk factors. Breast Cancer Res BCR 17:37. https://doi.org/10.1186/s13058-015-0546-7
Evans DG, Lalloo F, Cramer A, Jones EA, Knox F, Amir E, Howell A (2009) Addition of pathology and biomarker information significantly improves the performance of the Manchester scoring system for BRCA1 and BRCA2 testing. J Med Genet 46(12):811–817. https://doi.org/10.1136/jmg.2009.067850
Masciari S, Larsson N, Senz J, Boyd N, Kaurah P, Kandel MJ, Harris LN, Pinheiro HC, Troussard A, Miron P, Tung N, Oliveira C, Collins L, Schnitt S, Garber JE, Huntsman D (2007) Germline E-cadherin mutations in familial lobular breast cancer. J Med Genet 44(11):726–731. https://doi.org/10.1136/jmg.2007.051268
Benusiglio PR, Malka D, Rouleau E, De Pauw A, Buecher B, Nogues C, Fourme E, Colas C, Coulet F, Warcoin M, Grandjouan S, Sezeur A, Laurent-Puig P, Moliere D, Tlemsani C, Di Maria M, Byrde V, Delaloge S, Blayau M, Caron O (2013) CDH1 germline mutations and the hereditary diffuse gastric and lobular breast cancer syndrome: a multicentre study. J Med Genet 50(7):486–489. https://doi.org/10.1136/jmedgenet-2012-101472
Petridis C, Shinomiya I, Kohut K, Gorman P, Caneppele M, Shah V, Troy M, Pinder SE, Hanby A, Tomlinson I, Trembath RC, Roylance R, Simpson MA, Sawyer EJ (2014) Germline CDH1 mutations in bilateral lobular carcinoma in situ. Br J Cancer 110(4):1053–1057. https://doi.org/10.1038/bjc.2013.792
Corso G, Intra M, Trentin C, Veronesi P, Galimberti V (2016) CDH1 germline mutations and hereditary lobular breast cancer. Fam Cancer 15(2):215–219. https://doi.org/10.1007/s10689-016-9869-5
Pharoah PD, Guilford P, Caldas C, International Gastric Cancer Linkage C (2001) Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology 121(6):1348–1353. https://doi.org/10.1053/gast.2001.29611
Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, Schrader KA, Schaeffer DF, Shumansky K, Zogopoulos G, Santos TA, Claro I, Carvalho J, Nielsen C, Padilla S, Lum A, Talhouk A, Baker-Lange K, Richardson S, Lewis I, Lindor NM, Pennell E, MacMillan A, Fernandez B, Keller G, Lynch H, Shah SP, Guilford P, Gallinger S, Corso G, Roviello F, Caldas C, Oliveira C, Pharoah PD, Huntsman DG (2015) Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol 1(1):23–32. https://doi.org/10.1001/jamaoncol.2014.168
Ahmed M, Lalloo F, Evans DG (2009) Update on genetic predisposition to breast cancer. Expert Rev Anticancer Ther 9(8):1103–1113. https://doi.org/10.1586/era.09.38
Evans DG, Astley S, Stavrinos P, Harkness E, Donnelly LS, Dawe S, Jacob I, Harvie M, Cuzick J, Brentnall A, Wilson M, Harrison F, Payne K, Howell A (2016). In: Improvement in risk prediction, early detection and prevention of breast cancer in the NHS Breast Screening Programme and family history clinics: a dual cohort study. Programme Grants for Applied Research. Southampton (UK). https://doi.org/10.3310/pgfar04110
Breast Cancer Association C, Dorling L, Carvalho S, Allen J, Gonzalez-Neira A, Luccarini C, Wahlstrom C, Pooley KA, Parsons MT, Fortuno C, Wang Q, Bolla MK, Dennis J, Keeman R, Alonso MR, Alvarez N, Herraez B, Fernandez V, Nunez-Torres R, Osorio A, Valcich J, Li M, Torngren T, Harrington PA, Baynes C, Conroy DM, Decker B, Fachal L, Mavaddat N, Ahearn T, Aittomaki K, Antonenkova NN, Arnold N, Arveux P, Ausems M, Auvinen P, Becher H, Beckmann MW, Behrens S, Bermisheva M, Bialkowska K, Blomqvist C, Bogdanova NV, Bogdanova-Markov N, Bojesen SE, Bonanni B, Borresen-Dale AL, Brauch H, Bremer M, Briceno I, Bruning T, Burwinkel B, Cameron DA, Camp NJ, Campbell A, Carracedo A, Castelao JE, Cessna MH, Chanock SJ, Christiansen H, Collee JM, Cordina-Duverger E, Cornelissen S, Czene K, Dork T, Ekici AB, Engel C, Eriksson M, Fasching PA, Figueroa J, Flyger H, Forsti A, Gabrielson M, Gago-Dominguez M, Georgoulias V, Gil F, Giles GG, Glendon G, Garcia EBG, Alnaes GIG, Guenel P, Hadjisavvas A, Haeberle L, Hahnen E, Hall P, Hamann U, Harkness EF, Hartikainen JM, Hartman M, He W, Heemskerk-Gerritsen BAM, Hillemanns P, Hogervorst FBL, Hollestelle A, Ho WK, Hooning MJ, Howell A, Humphreys K, Idris F, Jakubowska A, Jung A, Kapoor PM, Kerin MJ, Khusnutdinova E, Kim SW, Ko YD, Kosma VM, Kristensen VN, Kyriacou K, Lakeman IMM, Lee JW, Lee MH, Li J, Lindblom A, Lo WY, Loizidou MA, Lophatananon A, Lubinski J, MacInnis RJ, Madsen MJ, Mannermaa A, Manoochehri M, Manoukian S, Margolin S, Martinez ME, Maurer T, Mavroudis D, McLean C, Meindl A, Mensenkamp AR, Michailidou K, Miller N, Mohd Taib NA, Muir K, Mulligan AM, Nevanlinna H, Newman WG, Nordestgaard BG, Ng PS, Oosterwijk JC, Park SK, Park-Simon TW, Perez JIA, Peterlongo P, Porteous DJ, Prajzendanc K, Prokofyeva D, Radice P, Rashid MU, Rhenius V, Rookus MA, Rudiger T, Saloustros E, Sawyer EJ, Schmutzler RK, Schneeweiss A, Schurmann P, Shah M, Sohn C, Southey MC, Surowy H, Suvanto M, Thanasitthichai S, Tomlinson I, Torres D, Truong T, Tzardi M, Valova Y, van Asperen CJ, Van Dam RM, van den Ouweland AMW, van der Kolk LE, van Veen EM, Wendt C, Williams JA, Yang XR, Yoon SY, Zamora MP, Evans DG, de la Hoya M, Simard J, Antoniou AC, Borg A, Andrulis IL, Chang-Claude J, Garcia-Closas M, Chenevix-Trench G, Milne RL, Pharoah PDP, Schmidt MK, Spurdle AB, Vreeswijk MPG, Benitez J, Dunning AM, Kvist A, Teo SH, Devilee P, Easton DF (2021) Breast cancer risk genes – association analysis in more than 113,000 women. N Engl J Med. https://doi.org/10.1056/NEJMoa1913948
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Moran A, O’Hara C, Khan S, Shack L, Woodward E, Maher ER, Lalloo F, Evans DG (2012) Risk of cancer other than breast or ovarian in individuals with BRCA1 and BRCA2 mutations. Fam Cancer 11(2):235–242. https://doi.org/10.1007/s10689-011-9506-2
Evans DG, Harkness EF, Plaskocinska I, Wallace AJ, Clancy T, Woodward ER, Howell TA, Tischkowitz M, Lalloo F (2017) Pathology update to the Manchester Scoring System based on testing in over 4000 families. J Med Genet 54(10):674–681. https://doi.org/10.1136/jmedgenet-2017-104584
Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, Devilee P, Meindl A, Couch FJ, Southey M, Goldgar DE, Evans DG, Chenevix-Trench G, Rahman N, Robson M, Domchek SM, Foulkes WD (2015) Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 372(23):2243–2257. https://doi.org/10.1056/NEJMsr1501341
Newman WG HK, McBurney H, Searle S, Lalloo F, Evans DG (2011) E-Cadherin (CDH1) exon deletion in lobular breast carcinoma. J Med Genet 48 (1)
Lu HM, Li S, Black MH, Lee S, Hoiness R, Wu S, Mu W, Huether R, Chen J, Sridhar S, Tian Y, McFarland R, Dolinsky J, Tippin Davis B, Mexal S, Dunlop C, Elliott A (2019) Association of breast and ovarian cancers with predisposition genes identified by large-scale sequencing. JAMA Oncol 5(1):51–57. https://doi.org/10.1001/jamaoncol.2018.2956
Weigelt B, Bi R, Kumar R, Blecua P, Mandelker DL, Geyer FC, Pareja F, James PA, kConFab I, Couch FJ, Eccles DM, Blows F, Pharoah P, Li A, Selenica P, Lim RS, Jayakumaran G, Waddell N, Shen R, Norton L, Wen HY, Powell SN, Riaz N, Robson ME, Reis-Filho JS, Chenevix-Trench G (2018) The landscape of somatic genetic alterations in breast cancers from ATM germline mutation carriers. J Natl Cancer Inst 110(9):1030–1034. https://doi.org/10.1093/jnci/djy028
Renault AL, Mebirouk N, Fuhrmann L, Bataillon G, Cavaciuti E, Le Gal D, Girard E, Popova T, La Rosa P, Beauvallet J, Eon-Marchais S, Dondon MG, d’Enghien CD, Lauge A, Chemlali W, Raynal V, Labbe M, Bieche I, Baulande S, Bay JO, Berthet P, Caron O, Buecher B, Faivre L, Fresnay M, Gauthier-Villars M, Gesta P, Janin N, Lejeune S, Maugard C, Moutton S, Venat-Bouvet L, Zattara H, Fricker JP, Gladieff L, Coupier I, Co FA, kConFab I, Chenevix-Trench G, Hall J, Vincent-Salomon A, Stoppa-Lyonnet D, Andrieu N, Lesueur F (2018) Morphology and genomic hallmarks of breast tumours developed by ATM deleterious variant carriers. Breast Cancer Res BCR 20(1):28. https://doi.org/10.1186/s13058-018-0951-9
Christgen M, Steinemann D, Kuhnle E, Langer F, Gluz O, Harbeck N, Kreipe H (2016) Lobular breast cancer: clinical, molecular and morphological characteristics. Pathol Res Pract 212(7):583–597. https://doi.org/10.1016/j.prp.2016.05.002
Yadav S, Hu C, Hart SN, Boddicker N, Polley EC, Na J, Gnanaolivu R, Lee KY, Lindstrom T, Armasu S, Fitz-Gibbon P, Ghosh K, Stan DL, Pruthi S, Neal L, Sandhu N, Rhodes DJ, Klassen C, Peethambaram PP, Haddad TC, Olson JE, Hoskin TL, Goetz MP, Domchek SM, Boughey JC, Ruddy KJ, Couch FJ (2020) Evaluation of germline genetic testing criteria in a hospital-based series of women with breast cancer. J Clin Oncol. https://doi.org/10.1200/JCO.19.02190
Blanco AM, Topper S, Nussbaum RL (2019) Association between invasive lobular breast cancer and mutations in the mismatch repair gene MSH6. JAMA Oncol 5(1):120–121. https://doi.org/10.1001/jamaoncol.2018.6911
Evans DG, Moran A, Hartley R, Dawson J, Bulman B, Knox F, Howell A, Lalloo F (2010) Long-term outcomes of breast cancer in women aged 30 years or younger, based on family history, pathology and BRCA1/BRCA2/TP53 status. Br J Cancer 102(7):1091–1098. https://doi.org/10.1038/sj.bjc.6605606
Smith MJ, Urquhart JE, Harkness EF, Miles EK, Bowers NL, Byers HJ, Bulman M, Gokhale C, Wallace AJ, Newman WG, Evans DG (2016) The contribution of whole gene deletions and large rearrangements to the mutation spectrum in inherited tumor predisposing syndromes. Hum Mutat 37(3):250–256. https://doi.org/10.1002/humu.22938
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Genome Aggregation Database C, Neale BM, Daly MJ, MacArthur DG (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581(7809):434–443. https://doi.org/10.1038/s41586-020-2308-7
Manickam K, Buchanan AH, Schwartz MLB, Hallquist MLG, Williams JL, Rahm AK, Rocha H, Savatt JM, Evans AE, Butry LM, Lazzeri AL, Lindbuchler DM, Flansburg CN, Leeming R, Vogel VG, Lebo MS, Mason-Suares HM, Hoskinson DC, Abul-Husn NS, Dewey FE, Overton JD, Reid JG, Baras A, Willard HF, McCormick CZ, Krishnamurthy SB, Hartzel DN, Kost KA, Lavage DR, Sturm AC, Frisbie LR, Person TN, Metpally RP, Giovanni MA, Lowry LE, Leader JB, Ritchie MD, Carey DJ, Justice AE, Kirchner HL, Faucett WA, Williams MS, Ledbetter DH, Murray MF (2018) Exome sequencing-based screening for BRCA1/2 expected pathogenic variants among adult biobank participants. JAMA Netw Open 1(5):e182140. https://doi.org/10.1001/jamanetworkopen.2018.2140
Acknowledgements
We thank the BRIDGES study investigators for their contribution to panel sequencing of a subset of our study participants. DGE, EFH, ERW, MJS, EMVV, HJB, WGN and SJH are supported by the Manchester NIHR Biomedical Research Centre (IS-BRC-1215-20007). JME is funded by a postdoctoral research fellowship from the Health Education England Genomics Education Programme (HEE GEP). The views expressed in this publication are those of the authors and not necessarily those of the HEE GEP.The genotyping work was supported by the Biomedical Research Centre and Prevent Breast Cancer (GA19-002).
Funding
The genotyping work was supported by the Biomedical Research Centre (IS-BRC-1215–20007) and Prevent Breast Cancer (GA19-002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
DGE has received travel grants from AstraZeneca. All other authors declare that they have no conflict of interest.
Ethical approval
Clinical or research consent was given for extended testing of breast cancer associated genes (approval from the North Manchester Research Ethics Committee (reference 09/H1008/81(PROCAS) and 08/H1006/77)).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van Veen, E.M., Evans, D.G., Harkness, E.F. et al. Extended gene panel testing in lobular breast cancer. Familial Cancer 21, 129–136 (2022). https://doi.org/10.1007/s10689-021-00241-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-021-00241-5