Abstract
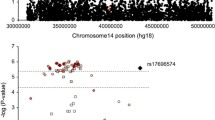
We aimed to detect Attention-deficit/hyperactivity (ADHD) risk-conferring genes in adults. In children, ADHD is characterized by age-inappropriate levels of inattention and/or hyperactivity-impulsivity and may persists into adulthood. Childhood and adulthood ADHD are heritable, and are thought to represent the clinical extreme of a continuous distribution of ADHD symptoms in the general population. We aimed to leverage the power of studies of quantitative ADHD symptoms in adults who were genotyped. Within the SAGA (Study of ADHD trait genetics in adults) consortium, we estimated the single nucleotide polymorphism (SNP)-based heritability of quantitative self-reported ADHD symptoms and carried out a genome-wide association meta-analysis in nine adult population-based and case-only cohorts of adults. A total of n = 14,689 individuals were included. In two of the SAGA cohorts we found a significant SNP-based heritability for self-rated ADHD symptom scores of respectively 15% (n = 3656) and 30% (n = 1841). The top hit of the genome-wide meta-analysis (SNP rs12661753; p-value = 3.02 × 10−7) was present in the long non-coding RNA gene STXBP5-AS1. This association was also observed in a meta-analysis of childhood ADHD symptom scores in eight population-based pediatric cohorts from the Early Genetics and Lifecourse Epidemiology (EAGLE) ADHD consortium (n = 14,776). Genome-wide meta-analysis of the SAGA and EAGLE data (n = 29,465) increased the strength of the association with the SNP rs12661753. In human HEK293 cells, expression of STXBP5-AS1 enhanced the expression of a reporter construct of STXBP5, a gene known to be involved in “SNAP” (Soluble NSF attachment protein) Receptor” (SNARE) complex formation. In mouse strains featuring different levels of impulsivity, transcript levels in the prefrontal cortex of the mouse ortholog Gm28905 strongly correlated negatively with motor impulsivity as measured in the five choice serial reaction time task (r2 = − 0.61; p = 0.004). Our results are consistent with an effect of the STXBP5-AS1 gene on ADHD symptom scores distribution and point to a possible biological mechanism, other than antisense RNA inhibition, involved in ADHD-related impulsivity levels.





Similar content being viewed by others
References
Achenbach TM, Dumenci L, Rescorla LA (2003) DSM-oriented and empirically based approaches to constructing scales from the same item pools. J Clin Child Adolesc Psychol 32(3):328–340
Antonucci F, Corradini I, Fossati G et al (2016) SNAP-25, a Known Presynaptic protein with emerging postsynaptic functions. Front Synaptic Neurosci 8:7
Aulchenko YS, Heutink P, Mackay I et al (2004) Linkage disequilibrium in young genetically isolated Dutch population. Eur J Hum Genet 12(7):527–534
Bonvicini C, Faraone SV, Scassellati C (2016) Attention-deficit hyperactivity disorder in adults: a systematic review and meta-analysis of genetic, pharmacogenetic and biochemical studies. Mol Psychiatr 21(7):872–884
Boomsma DI, Willemsen G, Sullivan PF et al (2008) Genome-wide association of major depression: description of samples for the GAIN Major depressive disorder study: NTR and NESDA biobank projects. Eur J Hum Genet 16(3):335–342
Bulik-Sullivan BK, Loh PR, Finucane HK et al (2015) LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47(3):291–295
Conners CK (1999) Clinical use of rating scales in diagnosis and treatment of attention-deficit/hyperactivity disorder. Pediatr Clin N Am 46(5):857–870, vi
Cupertino RB, Kappel DB, Bandeira CE et al (2016) SNARE complex in developmental psychiatry: neurotransmitter exocytosis and beyond. J Neural Transm 123(8):867–883
Davis LK, Meyer KJ, Rudd DS et al (2009) Novel copy number variants in children with autism and additional developmental anomalies. J Neurodev Disord 1(4):292–301
de Leeuw CA, Mooij JM, Heskes T et al (2015) MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11(4):e1004219
Demontis D, Walters RK, Martin J et al (2018) Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 51(1):63–75
Derks EM, Vink JM, Willemsen G et al (2014) Genetic and environmental influences on the relationship between adult ADHD symptoms and self-reported problem drinking in 6024 Dutch twins. Psychol Med 44(12):2673–2683
Dhillon S, Hellings JA, Butler MG (2011) Genetics and mitochondrial abnormalities in autism spectrum disorders: a review. Curr Genom 12(5):322–332
Distel MA, Carlier A, Middeldorp CM et al (2011) Borderline personality traits and adult attention-deficit hyperactivity disorder symptoms: a genetic analysis of comorbidity. Am J Med Genet B 156B(7):817–825
Faraone SV, Perlis RH, Doyle AE et al (2005) Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatr 57(11):1313–1323
Faraone SV, Asherson P, Banaschewski T et al (2015) Attention-deficit/hyperactivity disorder. Nat Rev Dis Primers 1:15020
Franke B, Faraone SV, Asherson P et al (2012) The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol Psychiatr 17(10):960–987
Franke B, Michelini G, Asherson P et al (2018) Live fast, die young? a review on the developmental trajectories of ADHD across the lifespan. Eur Neuropsychopharmacol 28(10):1059–1088
Galesloot TE, Vermeulen SH, Swinkels DW et al (2017) Cohort profile: the Nijmegen biomedical study (NBS). Int J Epidemiol 46(4):1099–1100j
Ghirardi L, Pettersson E, Taylor MJ et al (2018) Genetic and environmental contribution to the overlap between ADHD and ASD trait dimensions in young adults: a twin study. Psychol Med 7:1–9
Guillem K, Bloem B, Poorthuis RB et al (2011) Nicotinic acetylcholine receptor beta2 subunits in the medial prefrontal cortex control attention. Science 333(6044):888–891
Guo W, Wang Q, Zhan Y et al (2016) Transcriptome sequencing uncovers a three-long noncoding RNA signature in predicting breast cancer survival. Sci Rep 6:27931
Hoogman M, Rijpkema M, Janss L et al (2012) Current self-reported symptoms of attention deficit/hyperactivity disorder are associated with total brain volume in healthy adults. PLoS ONE 7(2):e31273
Kan KJ, Dolan CV, Nivard MG et al (2013) Genetic and environmental stability in attention problems across the lifespan: evidence from the Netherlands twin register. J Am Acad Child Adolesc Psychiatr 52(1):12–25
Kan KJ, van Beijsterveldt CE, Bartels M et al (2014) Assessing genetic influences on behavior: informant and context dependency as illustrated by the analysis of attention problems. Behav Genet 44(4):326–336
Kimura T, Jiang S, Nishizawa M et al (2013) Stabilization of human interferon-alpha1 mRNA by its antisense RNA. Cell Mol Life Sci 70(8):1451–1467
Klein M, Onnink M, van Donkelaar M et al (2017) Brain imaging genetics in ADHD and beyond—Mapping pathways from gene to disorder at different levels of complexity. Neurosci Biobehav Rev 80:115–155
Kooij JJ, Buitelaar JK, van den Oord EJ et al (2005) Internal and external validity of attention-deficit hyperactivity disorder in a population-based sample of adults. Psychol Med 35(6):817–827
Kuja-Halkola R, Lind Juto K, Skoglund C et al (2018) Do borderline personality disorder and attention-deficit/hyperactivity disorder co-aggregate in families? A population-based study of 2 million Swedes. Mol Psychiatr. https://doi.org/10.1038/s41380-018-0248-5.
Larsson H, Asherson P, Chang Z et al (2013) Genetic and environmental influences on adult attention deficit hyperactivity disorder symptoms: a large Swedish population-based study of twins. Psychol Med 43(1):197–207
Larsson H, Chang Z, D’Onofrio BM et al (2014) The heritability of clinically diagnosed attention deficit hyperactivity disorder across the lifespan. Psychol Med 44(10):2223–2229
Loos M, Mueller T, Gouwenberg Y et al (2014) Neuregulin-3 in the mouse medial prefrontal cortex regulates impulsive action. Biol Psychiatr 76(8):648–655
Matsui K, Nishizawa M, Ozaki T et al (2008) Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology 47(2):686–697
Middeldorp CM, Hammerschlag AR, Ouwens KG et al (2016) A genome-wide association meta-analysis of attention-deficit/hyperactivity disorder symptoms in population-based pediatric cohorts. J Am Acad Child Adolesc Psychiatr 55(10):896–905.e896
Mostert JC, Onnink AMH, Klein M et al (2015) Cognitive heterogeneity in adult attention deficit/hyperactivity disorder: a systematic analysis of neuropsychological measurements. Eur Neuropsychopharmacol 25(11):2062–2074
Oldehinkel AJ, Rosmalen JG, Buitelaar JK et al (2015) Cohort profile update: the tracking adolescents’ individual lives survey (TRAILS). Int J Epidemiol 44(1):76–76n
Pinto R, Monzani B, Leckman JF et al (2016) Understanding the covariation of tics, attention-deficit/hyperactivity, and obsessive-compulsive symptoms: a population-based adult twin study. Am J Med Genet B 171(7):938–947
Polanczyk G, Rohde LA (2007) Epidemiology of attention-deficit/hyperactivity disorder across the lifespan. Curr Opin Psychiatr 20(4):386–392
Psychiatric Genomics Consortium CDS, Lee SH, Ripke S et al (2013) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45(9):984–994
Richarte V, Corrales M, Pozuelo M et al (2017) Spanish validation of the adult attention deficit/hyperactivity disorder rating scale (ADHD-RS): relevance of clinical subtypes. Rev Psiquiatr Salud Ment 10(4):185–191
Sakisaka T, Baba T, Tanaka S et al (2004) Regulation of SNAREs by tomosyn and ROCK: implication in extension and retraction of neurites. J Cell Biol 166(1):17–25
Sakisaka T, Yamamoto Y, Mochida S et al (2008) Dual inhibition of SNARE complex formation by tomosyn ensures controlled neurotransmitter release. J Cell Biol 183(2):323–337
Spijker S, Houtzager SW, De Gunst MC et al (2004) Morphine exposure and abstinence define specific stages of gene expression in the rat nucleus accumbens. FASEB J 18(7):848–850
Thissen AJ, Rommelse NN, Altink ME et al (2012) Parent-of-origin effects in ADHD: distinct influences of paternal and maternal ADHD on neuropsychological functioning in offspring. J Atten Disord 18(6):521–531
von Rhein D, Mennes M, van Ewijk H et al (2014) The NeuroIMAGE study: a prospective phenotypic, cognitive, genetic and MRI study in children with attention-deficit/hyperactivity disorder. Design and descriptives. Eur Child Adolesc Psychiatr 24(3):265–281
Willemsen G, de Geus EJ, Bartels M et al (2010) The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res Hum Genet 13(3):231–245
Wu MC, Lee S, Cai T et al (2011) Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89(1):82–93
Yang J, Lee SH, Goddard ME et al (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1):76–82
Yizhar O, Matti U, Melamed R et al (2004) Tomosyn inhibits priming of large dense-core vesicles in a calcium-dependent manner. Proc Natl Acad Sci USA 101(8):2578–2583
Acknowledgements
This work was sponsored by the Stichting Nationale Computerfaciliteiten (National Computing Facilities Foundation, NCF) for the use of supercomputer facilities, with financial support from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organization for Scientific Research, NWO) This work made use of the BIG (Brain Imaging Genetics) database, first established in Nijmegen, The Netherlands, in 2007. This resource is now part of Cognomics (http://www.cognomics.nl), a joint initiative by researchers of the Donders Centre for Cognitive Neuroimaging, the Human Genetics and Cognitive Neuroscience departments of the Radboud University Medical Center and the Max Planck Institute for Psycholinguistics in Nijmegen. The Cognomics Initiative is supported by the participating departments and centres and by external grants, i.e. the Biobanking and Biomolecular Resources Research Infrastructure (Netherlands) (BBMRI-NL), the Hersenstichting Nederland, and the Netherlands Organisation for Scientific Research (NWO). We wish to thank all persons who kindly participated in the BIG research. The research leading to these results also receives funding from the European Community‘s Seventh Framework Programme (Grant No. FP7/2007–2013) under grant agreements #602450 (IMAGEMEND) and #602805 (Aggressotype), and from ERC-2010-AdG 268800-NEUROSCHEMA. In addition, we are funded by a grant from the National Institutes of Health (NIH) for the ENIGMA Consortium (Consortium grant U54 EB020403), supported by a cross-NIH alliance that funds big data to knowledge centers of excellence. The erasmus rucphen family study (ERF) study as a part of EUROSPAN (European Special Populations Research Network) was supported by European Commission FP6 STRP Grant Number 018947 (Grant No. LSHG-CT-2006-01947) and also received funding from the European Community’s Seventh Framework Program (Grant No. FP7/2007–2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the program ‘Quality of Life and Management of the Living Resources’ of 5th Framework Program (Grant No. QLG2-CT-2002-01254). High-throughput analysis of the ERF data was supported by joint grant from Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). Exome sequencing analysis in ERF was supported by the ZonMw grant (project 91111025). Najaf Amin is supported by the Hersenstichting Nederland (project number F2013(1)-28). We are grateful to all study participants and their relatives, general practitioners and neurologists for their contributions and to P Veraart for her help in genealogy, J Vergeer for the supervision of the laboratory work, P Snijders for his help in data collection and E.M. van Leeuwen for genetic imputation. The International Multicentre persistent ADHD CollaboraTion (IMpACT). IMpACT unites major research centres working on the genetics of ADHD persistence across the lifespan and has participants in the Netherlands, Germany, Spain, Norway, the United Kingdom, the United States, Brazil and Sweden. Principal investigators of IMpACT are: Barbara Franke (chair), Andreas Reif, Stephen V. Faraone, Jan Haavik, Bru Cormand, Antoni Ramos Quiroga, Philip Asherson, Klaus-Peter Lesch, Jonna Kuntsi, Claiton Bau, Jan Buitelaar, Stefan Johansson, Henrik Larsson, Alysa Doyle, and Eugenio Grevet. The Dutch IMpACT study is supported by grants from the Netherlands Organization for Scientific Research (NWO), i.e. the NWO Brain & Cognition Excellence Program (Grant No. 433-09-229) and a Vici grant to BF (Grant No. 016-130-669), and by grants from the Netherlands Brain Foundation (Grant No. 15F07[2]27) and BBMRI-NL (Grant No. CP2010-33). The research leading to these results also received funding from the European Community’s Seventh Framework Programme (Grant No. FP7/2007–2013) under grant agreements n° 602805 (Aggressotype), n° 278948 (TACTICS), and n° 602450 (IMAGEMEND), and from the European Community’s Horizon 2020 Programme (H2020/2014–2020) under grant agreement n° 643051 (MiND) and n° 667302 (CoCA). For the Netherlands Study of Depression and Anxiety (NESDA), funding was obtained from the Netherlands Organization for Scientific Research (Geestkracht program grant 10-000-1002); the Center for Medical Systems Biology (CSMB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL), VU University’s Institutes for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam, University Medical Center Groningen, Leiden University Medical Center, National Institutes of Health (NIH, R01D0042157-01A, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. Computing was supported by BiG Grid, the Dutch e-Science Grid, which is financially supported by NWO. The NeuroIMAGE study was supported by grants from The Netherlands Organization for Health Research and Development (ZonMw 60-60600-97-193), the Netherlands Organization for Scientific Research (NWO, grants 1750102007010, 433-09-242 and 056-13-015), and by the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement number 278948 (TACTICS), 602450 (IMAGEMEND), 602805 (AGGRESSOTYPE), 603016 (MATRICS), and Horizon 2020 (grant agreements 643051 (MiND) and 642996 (BRAINVIEW)) research programs. The Nijmegen Biomedical Study (NBS) is a population-based survey conducted at the Department for Health Evidence, and the Department of Laboratory Medicine of the Radboud University Medical Center. Principal investigators of the Nijmegen Biomedical Study are L.A.L.M. Kiemeney, A.L.M. Verbeek, D.W. Swinkels and B. Franke. NTR Research was funded by the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW grants 904-61-090, 985-10-002,904-61-193,480-04-004, 400-05-717, 91210020, Addiction-31160008, Middelgroot-911-09-032, Spinozapremie 56-464-14192 and NWO 480-15-001/674), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL, 184.021.007), and the European Research Council (230374 and 284167). We thank the Avera Institute of Human Genetics for genotyping and technical support. DIB acknowledges the Royal Netherlands Academy of Science Professor Award (PAH/6635) We thank all participants in the Netherlands Twin Register. The TRacking Adolescents’ Individual Lives Survey (TRAILS) is a collaborative project involving various departments of the University Medical Center and University of Groningen, the Erasmus University Medical Center Rotterdam, the University of Utrecht, the Radboud Medical Center Nijmegen, and the Parnassia Bavo group, all in the Netherlands. TRAILS has been financially supported by grants from the Netherlands Organization for Scientific Research NWO (Medical Research Council program grant GB-MW 940-38-011; ZonMW Brainpower grant 100-001-004; ZonMw Risk Behavior and Dependence grant 60-60600-97-118; ZonMw Culture and Health grant 261-98-710; Social Sciences Council medium-sized investment grants GB-MaGW 480-01-006 and GB-MaGW 480-07-001; Social Sciences Council project grants GB-MaGW 452-04-314 and GB-MaGW 452-06-004; NWO large-sized investment grant 175.010.2003.005; NWO Longitudinal Survey and Panel Funding 481-08-013); the Dutch Ministry of Justice (WODC), the European Science Foundation (EuroSTRESS project FP-006), Biobanking and Biomolecular Resources Research Infrastructure BBMRI-NL (CP 32), the participating universities, and Accare Center for Child and Adolescent Psychiatry. We are grateful to all adolescents, their parents and teachers who participated in this research and to everyone who worked on this project and made it possible. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (Grant No. NWO 480-05-003) along with a supplement from the Dutch Brain Foundation. The Vall d’Hebron Research Institute (VHIR) received financial support from “Fundació La Marató de TV3” (ref. 092330/31), “Instituto de Salud Carlos III-FIS” (Grant Nos. PI11/00571, PI11/01629, PI12/01139, PI14/01700), “Agència de Gestió d’Ajuts Universitaris i de Recerca-AGAUR, Generalitat de Catalunya” (Grant No. 2014SGR1357) and “Departament de Salut”, Government of Catalonia, Spain. We wish to thank all persons who kindly participated in this research. Part of the DNA extractions and genotyping was performed at the Spanish National Genotyping Centre (CEGEN-Barcelona). Harmen H.M. Draisma is supported by an EMGO + Fellowship as part of the Mental Health research program of the EMGO Institute for Health and Care Research. Sabine Spijker was partially funded by the Center for Medical Systems Biology (CMSB). Sander Goffen was financially supported by grant 91113022 from the Netherlands Organization for Health Research and Development (ZonMW). Marta Ribasés is a recipient of a Miguel de Servet contract from the “Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación”, Spain. Barbara Franke’s research is supported by grants from the Netherlands Organisation for Scientific Research (NWO; grants 433-09-229 and 016-130-669), from the European Union 7th Framework (grant agreements 278948 (TACTICS), 602450 (IMAGEMEND), and 602805 (Aggressotype)) and Horizon 2020 (grant agreements 643051 (MiND) and 667302 (CoCA)) research programmes. A.B. Smit was partially funded by the Dutch Neuro-Bsik Mouse Pharma Phenomics consortium (grant BSIK 03053 from SenterNovem). This work was partly carried out on the Dutch national e-infrastructure with the support of SURF Foundation.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
J.K.B. has been in the past 3 years a consultant to/member of advisory board of/and/or speaker for Janssen Cilag BV, Eli Lilly, Shire, Lundbeck, Roche and Servier. He is not an employee of any of these companies, and not a stock shareholder of any of these companies. He has no other financial or material support, including expert testimony, patents, royalties. J.J.S.K. has been a speaker for Eli Lilly, Janssen and Shire until 2012, and received unrestricted research grants in 2010 from Janssen and Shire. J.A.R.Q. was on the speakers’ bureau and/or acted as consultant for Eli-Lilly, Janssen-Cilag, Novartis, Shire, Lundbeck, Ferrer and Rubió in the last 3 years. He also received travel awards (air tickets + hotel) for taking part in psychiatric meetings from Janssen-Cilag, Rubió, Shire and Eli-Lilly. The ADHD Program chaired by him received unrestricted educational and research support from the following pharmaceutical companies in the last 3 years: Eli-Lilly, Janssen-Cilag, Shire, Rovi and Rubió. B.F. received educational speaking fees from Shire and Medice. A.A.V., A.J.G., H.H.M.D., M.K., D.V., S.S., T.E.G., J.J.H., P.J.vdM., V.M. K, R.P., I.M.N., B.W.J.H.P., I.O.F, A.dB., C.M.vD., P.J.H., L.A.K., M.H, M.K., C.M.M., K.G.O., S.H.V., C.S.M., M.R., C.A.H., N.A., A.B.S., D.I.B. report no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Stacey Cherny.
A list of members and affiliations appears in the Supplementary Notes.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Arias-Vásquez, A., Groffen, A.J., Spijker, S. et al. A Potential Role for the STXBP5-AS1 Gene in Adult ADHD Symptoms. Behav Genet 49, 270–285 (2019). https://doi.org/10.1007/s10519-018-09947-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-018-09947-2