ABSTRACT
Background
Congenital disorders of glycosylation (CDG) includes ALG8 deficiency, a protein N-glycosylation defect with a broad clinical spectrum. If most of the 15 previously reported patients present an early-onset multisystem severe disease and early death, three patients including the cas princeps, present long-term survival and less severe symptoms.
Methods
In order to further characterize ALG8-CDG, two new ALG8 patients are described and mRNA analyses of the ALG8-CDG cas princeps were effected.
Results
One new patient exhibited a hepato-intestinal and neurological phenotype with two novel variants (c.91A > C p.Thr31Pro; c.139dup p.Thr47Asnfs*12). The other new patient, homozygous for a known variant (c.845C > T p.Ala282Val), presented a neurological phenotype with epilepsy, intellectual disability and retinis pigmentosa. The cas princeps ALG8-CDG patient was reported to have two heterozygous frameshift variants predicted to be without activity. We now described a novel ALG8 transcript variant in this patient and the 3D model of the putative encoded protein reveals no major difference with that of the normal ALG8 protein.
Conclusion
The description of the two new ALG8 patients affirms that ALG8-CDG is a severe disease. In the cas princeps, as the originally described frameshift variants are degraded, the novel variant is promoted and could explain a milder phenotype.
Similar content being viewed by others
Introduction
Impaired N- or O-glycosylation of proteins and lipids causes a family of diseases called congenital disorders of glycosylation (CDG).1,2 Historically, CDG were classified by transferrin isoform analysis. CDG-I comprises alterations in the assembly of the lipid-linked oligosaccharide (LLO) N-glycosylation precursor or the transfer of its oligosaccharide moiety onto proteins in the endoplasmic reticulum. CDG-II concerns the remodeling defects of glycans in the Golgi apparatus. More than 100 inborn errors of N-linked protein glycosylation metabolism have been described.2,3,4 CDG-I are generally multisystem disorders with a wide variety of severe to mild clinical presentations, and most often present with nervous system deficits. Mutations in the PMM2 gene cause the most frequent type of CDG: PMM2-CDG, which occurs as a neurological form or a more severe neurovisceral form.5 Mutations in the ALG8 gene lead to ALG8-CDG (OMIM #608104).6 This gene encodes the dolichyl-P-glucose:Glc1Man9GlcNAc2-PP-dolichyl α3-glucosyltransferase that adds the second glucose residue onto LLO. Man9GlcNAc2- and Glc1Man9GlcNAc2-containing LLO intermediates accumulate in fibroblasts leading to a block in LLO assembly and glycosylation deficiency.6
In all, 15 patients from 11 families have been described with ALG8-CDG6,7,8,9,10,11,12 and published data concerning these cases have been reviewed.13 By contrast to the first patient to be described as ALG8-CDG by Chantret et al.,6 currently 18 years old, 12 ALG8-CDG patients died between 15 min and 4 years of life, 3 of which had prenatal onset with intrauterine growth retardation, reduced fetal movement and/or oligohydramnios.9,10
Of the three patients presenting with long-term survival, the first one6 had normal early psychomotor development until 6 years of age when she started requiring help and subsequently specialized schooling. She exhibited early-onset protein-losing enteropathy with severe diarrhea, hypoalbuminemia, and moderate hepatomegaly and ascites. The other two surviving patients, German siblings described by Stolting et al.,11 presented with a mild central nervous system phenotype (muscular hypotonia, mental retardation, and ataxia).
Here we provide long-term follow-up data on the first-described ALG8-CDG patient6 (P1), and report on two other previously unreported ALG8-CDG patients, an 11-year-old patient (P2) presenting with epilepsy, intellectual disability (ID), and retinitis pigmentosa, and a 10-month patient (P3), with protein-losing enteropathy, cardiac involvement, hepatomegaly, and neurological impairment. We investigated the molecular origin of the particularly mild phenotype of ALG8-CDG observed in P1.
Patients
Informed consent was obtained from all patients included in the study. Findings are summarized in Table 1.
Patient 1 (P1)
A girl born to unrelated French healthy parents, was referred at 4 months of age for an edemato-ascitic syndrome related to severe hypoalbuminemia (8 g/L, normal value >30 g/L) resulting from protein-losing enteropathy. She exhibited severe diarrhea, and moderate hepatomegaly but normal initial psychomotor development and no dysmorphic signs.
She was treated with a low-fat diet with essential fatty acid supplementation. Coagulation factors were persistently abnormal with an activated cephalin time (ACT) of 51 s (normal value: <30 s) and factor XI at 27% (normal range (NR): 80–120).
At 5 years of age, she exhibited a relapse of hypoalbuminemia at 12 g/L requiring albumin infusion. Five months later, serum proteins were 51 g/L and albumin 28.3 g/L, coagulation factors were abnormal with ACT at 57 s, antithrombin at 42% (NR: 70–120%), C protein at 55% (NR: 70–120%). Intestinal symptoms recovered with albumin infusions and parenteral nutrition followed by an oral low-fat diet consisting of 15% energy intake in the form of lipids supplemented with essential fatty acids. Chronic hypoalbuminemia and mild protein-losing enteropathy without diarrhea have persisted. From time to time, palpebral edema was noticed but she had required only a few albumin infusions in infancy. Hepatomegaly (with normal serum transaminases and liver ultrasound) is still present. Now, at 18 years, she presents schooling difficulties due to dyspraxia and ID (intelligence quotient at 71, WISC-IV (Wechsler Intelligence Scale for Children-IV)) requiring specialized education that started around the age of 6 years. Brain magnetic resonance imaging (MRI) is normal. With mild lipid dietary restriction, she has had no relapse of hypoalbuminemia. Electroretinogram is normal.
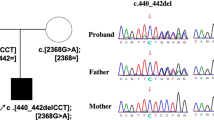
Diagnosis of CDG-I was made upon investigation of the glycosylation status of the serum glycoproteins by Western blot when she was 5 months. Radiolabeling of fibroblast LLO with [3H]mannose demonstrated an accumulation of Man9GlcNAc2. After addition of castanospermine, an endoplasmic reticulum glucosidase inhibitor, Glc1Man9GlcNAc2 predominated, suggesting inefficient addition of the second glucose residue onto LLO. Genomic sequencing of ALG8 revealed compound heterozygosity for two frameshift mutations: one deletion of cytosine from paternal allele c.413delC p.(Thr138Lysfs*19) and one insertion of adenine from maternal allele c.396dup p.(Val133Serfs*3), both in exon 4.6
Patient 2 (P2)
The second patient is an 11-year-old boy, born of non-consanguineous (but from the same village) parents of Turkish ancestry. Dysmorphic signs were noted: bitemporal retraction with horizontal eyebrows, enophthalmia, slightly short and tapered fingers, and hypogenitalism. At 9 months of age, he was hospitalized for seizures and treated with carbamazepine. The absence of recurrence allowed the treatment to be stopped at 5 years. He walked at 2 years of age with frequent falls, and presented speech delay. He required specialized education and multidisciplinary care. At present, his language is limited to a few words, reading and writing have not been acquired, and comprehension is limited to simple orders.
Obesity was diagnosed in the first years of life and was difficult to control as the patient presents hyperphagia and aggressive behavior.
The patient has had regular ophthalmological evaluations. At 8 years, bilateral retinitis pigmentosa was diagnosed, with low vision (electroretinographic was not performed due to behavioral troubles).
Recently, epilepsy, characterized by generalized tonico–clonic seizures during sleep, was diagnosed. A treatment with topiramate and clabazam was initiated. The neurological evolution was marked by regressive episodes with hypotonia and extreme tiredness. Extensive medical checks found no anomaly (the computed tomography scan was notably normal). The last neurological examination showed ataxia, myoclonia and action tremors. The deep tendon reflexes were exaggerated but without pyramidal signs. Liver size was normal. Laboratory tests showed hypoalbuminemia (35 g/L (N > 39 g/L), and abnormal coagulation factors with factor XI at 55% (NR: 60–120%), protein C at 60% (NR: 70–120%) and AT at 55% (NR: 80–120%).
Several genetic analyses were performed (karyotype, Fragile X syndrome, Angelmann syndrome, Prader–Willi syndrome, UBE3A sequencing, array comparative genomic hybridization, high-throughput sequencing of 225 genes involved in ID) but no pathogenic variants were found. Whole-exome sequencing revealed a homozygous missense variant in exon 8 of the ALG8 gene: c.845C > T, p.(Ala282Val). This was confirmed by Sanger sequencing. Hypoglycosylation of serum glycoproteins was found subsequently.
Patient 3 (P3)
This 10-month-old boy, born eutrophic from non-consanguineous French parents, exhibited early-onset (from 3 months of age) hypotonia and was admitted for subacute neurological deterioration with exudative enteropathy with ascites, hepatomegaly, hypoalbuminemia at 13 g/L, and a transient prolonged QT interval (EKG). Factor XI and AT were at 29 and 23%, respectively. Prothrombin time and factor V were normal. After 2 weeks of albumin infusions the patient recovered. Generalized epilepsy occurred at 15 months and was well controlled by Levetiracetam. At the latest examination, the boy was 28 months of age and presented with neurodevelopmental disability, hypotonia, and mild hepatomegaly. His weight was 11.6 kg (−0.5 SD), height 85 cm (−0.5 SD), and head circumference 46 cm (−1 SD). Brain MRI, performed at 10 months, showed moderate white matter atrophy without cerebellar hypoplasia.
Diagnosis of ALG8-CDG was made after detecting hypoglycosylation of serum glycoproteins typical of CDG-I. PMM activity was normal, excluding PMM2-CDG, and targeted sequencing with a custom kit (Agilent) of 17 CDG-I genes revealed compound heterozygosity for two ALG8 mutations: one missense mutation inherited from the mother c.91A > C p.(Thr31Pro) in exon 1 and one frameshift inherited from the father c.139dupA p.(Thr47Asnfs*12) on exon 2. Mutations and segregation were confirmed by Sanger sequencing.
Methods
Identification of the ALG8 alternative splicing for P1
RNA was isolated from EBV-transformed lymphoblasts derived from P1 and fresh blood cells from the parents withdrawn into “Paxgene tubes” (Qiagen, USA). In order to abolish mRNA degradation caused by translation-dependent nonsense-mediated mRNA decay (NMD), we treated the cells from the patient with the translation inhibitor emetine at the concentration of 10 µg/mL before RNA extraction, as previously described.14
Reverse transcription was performed on total RNA with Superscript II (Gibco BRL, CA, USA) and an oligodT primer. Primer sequences for PCR are given in Supplementary Table S1. The size of complementary DNA-generated amplicons, which have been obtained by PCR using the primers couple 2S–2AS, was studied on agarose gel. PCR products were subcloned into pCR3.1. After purification, direct sequencing was performed with the Big Dye terminator kit (Applied Biosystems, CA, USA) and sequences were analyzed on an ABI PRISM 3130 capillary sequencer (Applied Biosystems, CA, USA).
Pathogenicity prediction of ALG8 missense variants
The pathogenicity of the missense variants was evaluated in silico with five software programs:
-
(1)
Polyphen (http://genetics.bwh.harvard.edu/pph/).15
-
(2)
Panther (http://www.pantherdb.org/tools/csnpScoreForm.jsp).16
-
(3)
SIFTV2 (http://blocks.fhcrc.org/sift/SIFT_seq_submit2.html).17
- (4)
-
(5)
MutationTaster (http://www.mutationtaster.org/).19
Splice sites prediction of ALG8 exon 3
Potential splice sites and splice scores of ALG8 exon 3 were evaluated in silico using the Human Splicing Finder software program.20
3D modeling of ALG8
The online webserver I-TASSER (Iterative Threading ASSEmbly Refinement) was used to determine a 3D model of ALG8. Based on structural templates from the pdb by multiple threading alignment,21 indicating that ALG8 showed homology with the oligosaccharyltransferase from Archaeoglobus fulgidus (pdb: 3waj), 3D models corresponding to the wild-type (WT) and variant forms of the enzyme were constructed. The 3D models were energy minimized using Gromos96 vacuum force field.22 Models were visualized with USCF Chimera software.23
Results
The first French ALG8-CDG patient, P1, is affected with an exceptionally mild form of this disease. She is currently 18 years old with moderate ID and dyspraxia. She presented a high residual ALG8 expression, which appears to be inconsistent with compound heterozygosity for two supposed severe frameshift variants.6 She inherited an allele with one base deletion from the father (c.413delC), and a maternal allele with one base insertion (c.396dup). Both mutations are located in exon 4 and give rise to premature stop codons predicted to generate severely truncated proteins. The level of the ALG8 mRNA found in cells from the patient was found to be 10–20% of control level by Northern blot analysis.
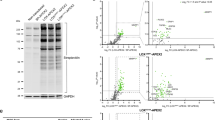
Sequencing of ALG8 mRNA in P1 identified a new transcript variant associated with the mutated maternal allele: this transcript had a deletion of four base pairs before the insertion of one base mutation, which allowed reestablishment of an open reading frame after a frameshift of seven amino acids from 123 to 129 (Fig. 1). The seven amino acid sequence is not found in any of the databases. Analysis by four software programs revealed that substitution of the new amino acids, one by one, into the WT sequence was well tolerated (Supplementary Table S2). This transcript variant may derive from activation of a cryptic splice site located at the four last bases of exon 3, which is of similar force (81.1) to that of the normal donor splice site (82.58) (Fig. 1). The modeling of WT and variant of ALG8 using I-TASSER multi-threading approach indicated that WT ALG8 had an RMSD (root mean square deviation) of 11.8 Å and variant had an RMSD of 11.3 Å, the amino acid substitution region structured as an α-helix, was located at the end of TM2 in the interface between membrane and the intra-endoplasmic reticulum compartment. The structural comparison between ALG8 WT and variant showed a very similar structure (RMSD = 3.22 Å), indicating that the amino acid substitution had no impact on the structure of ALG8 (Fig. 2). Furthermore, the newly described transcript variant was less frequently observed after treatment of P1 cells with emetine proving that this variant is neither a substrate for NMD nor an artifact of reverse transcription-PCR. The alternative spliced variant was quantified after subcloning and was found to correspond to 55.6% of the total transcript compared to 22.2% for each of the mutated alleles (Table 2). After emetine treatment, the variant transcript could not be detected, and it was apparent that the transcript originating from the father (71.4% of total) was more stabilized than that derived from the mother (28.6% of total).
Scheme showing activation of a cryptic splice site in exon 3–intron 3 of the ALG8 allele derived from the mother of patient P1. Position of the normal splice site between exon 3 and intron 3 of the normal ALG8 allele (N) is shown. In the maternal allele (M) of P1, a 1 bp insertion generates a premature termination codon (PTC), which leads to the translation of a highly truncated protein with no activity. In the new variant mRNA (V) found in P1, a cryptic splice site causes a 4 bp deletion from the end of exon 3 and allows recovery of the open reading frame at the position of the base insertion corresponding to the mother’s mutation in exon 4, but with the generation of a 7 amino acid sequence (indicated in red) that differs from the normal peptidic sequence. The splice scores (calculated with Human Splicing Finder software (HSF)) are indicated above the corresponding tandem donor site and are similar between cryptic site and normal donor splice site
The 3D modeling of protein encoded by the normal ALG8 mRNA (a) and of the protein encoded by the variant transcript in P1 (b). Based on the structural homology with the oligosaccharyltransferase from Archaeoglobus fulgidus (pdb: 3waj), we can delineate two domains in ALG8 protein: the transmembrane domain (below the dotted line) and the ER luminal domain (above the dotted line). Note that the 7 amino acid substitutions found in the peptide sequence deduced from the transcript variant (AVNALME instead of ECCKCID) had only minor impact on the structure of the ALG8 protein
P2 presented with epilepsy and ID and a homozygous missense variant c.845C > T p.(Ala282Val), which was predicted to be pathogenic by 4/5 prediction software tools (SIFT, MutationTaster, Panther, and Polyphen2) and has been previously described in the two German patients with mild disease described by Stolting et al.11
P3, presented with the more frequently observed early-onset hepato-intestinal and neurological phenotype with the c.91A > C p.Thr31Pro and c.139dupA p.(Thr47Asnfs*12) heterozygous variants. The missense mutation p.(Thr31Pro) was predicted pathogenic by 3/5 prediction software tools (Polyphen2, Panther, and MutationTaster) out of 5.
Discussion
CDG presents a great diversity of clinical presentations even within the same subtype. This is well illustrated by the three ALG8-CDG patients presented here (Table 1). Common findings were hypoalbuminemia, hepatomegaly, and abnormal coagulation factors, which could suggest a CDG syndrome in these three patients, but frequent CDG dysmorphic features such as inverted nipples and abnormal fat distribution present in around 50% of patients were not present, as well as low-set ears, hypertelorism, and macroglossia, which have also been described in ALG8-CDG.4 Patient P1 had no neurological signs at birth, no dysmorphic features, and presented a high residual ALG8 expression inconsistent with compound heterozygosity for two frameshift variants. The clinical course obviously differs from the other reported cases except for two German siblings described by Stolting et al.11 presenting also with a mild form. It is well known that mRNA containing premature stop codons can be degraded by NMD, a quality control mechanism that selectively degrades mRNA harboring premature termination codons (PTCs); if translated, these mRNAs can produce truncated proteins with dominant-negative or deleterious gain-of-function activities. NMD prevents the accumulation of truncated proteins in genetic disease: approximately one-third of all known disease-associated mutations result in the generation of mRNAs with a PTC.24
We have been able to show that both ALG8 transcripts of P1 undergo NMD because the translation inhibitor emetine stabilizes their mRNA levels. However, the paternal transcript was more stabilized than the maternal allele (Table 2). We believe that we were unable to detect the variant transcript under these conditions because: first, it is not stabilized by emetine, and second, the other transcripts become dominant.
Despite the very low level of ALG8 mRNA, non-negligible dolichyl-P-glucose:Glc1Man9GlcNAc2-PP-dolichylα3-glucosyltransferase activity was observed as lipid-linked Glc2Man9GlcNAc2and Glc3Man9GlcNAc2 structures were found as well as Man9GlcNAc2 and Glc1Man9GlcNAc2.6 A possible explanation could be that the maternal mutation in exon 4 creates a silencer octamer motif of eight adenines that could be in favor of alternative splicing.20 This kind of alternative splicing at donor or acceptor sites located just a few nucleotides apart is widespread in many species and contributes to transcriptome and proteome diversity.25 Such splice site pairs are so-called tandem sites.26 One-third of all alternative donor and acceptor pairs are located 2–10 nucleotides apart.27 At the donor site, a nucleotide distance of 4 is more frequent because the intronic donor consensus sequence GTRAGT (R = A, G) provides a second GT 4 nucleotides from the donor site in 40% of introns.28 This is the case for the new ALG8 variant, which is generated by a cryptic exonic splice site, located 4 bases upstream of the usual exon 3 splice donor site (Fig. 1).
This new ALG8 transcript variant could explain the recovery of residual ALG8 expression from the mutated maternal allele and could explain why the clinical symptoms are not so severe in the first-described ALG8-CDG patient (P1). It is known that alternative splicing coupled with NMD efficiently regulates gene expression.29 Moreover, the modeling of WT and the variant ALG8 shows that the amino acid substitutions do not strikingly modify the structure of the protein explaining the absence of severe effect of this mutation (Fig. 2).
For the two German siblings described by Stolting et al.,11 a small amount of mature LLO (Glc3Man9GlcNAc2-PP-dolichol) and a mild phenotype were also observed. In these patients, alternative splicing has also been proposed to alleviate the consequences of a deleterious mutation in a highly conserved region and 25% of the transcripts would encoded for a protein which should have an ALG8 activity.
Patient P3, who has a severe phenotype harbors compound heterozygosity of severe mutations: one missense mutation predicted to be pathogenic c.91A > C p.(Thr31Pro) in exon 1 and one frameshift c.139dupA p.(Thr47Asnfs*12) mutation in exon 2. The latter mutation has already been reported, in association with a severe missense mutation p.(Thr147Pro), in seven other severely affected ALG8-CDG patients.13
To conclude, these results demonstrate the continuous spectrum of phenotypes caused by mutations in the ALG8 gene and suggest that the severity of the disease may depend upon the presence or absence of mechanisms such as alternative splicing and NMD that can modulate the consequence of the damaging mutation. While 15 years ago, CDG diagnosis was initially performed with biochemical tools and then confirmed by molecular data (P1), currently, whole-exome sequencing allows diagnosis of patients whose clinical presentation is not suggestive of CDG (P2) and allows to widen the clinical spectrum of ALG8-CDG.
References
Jaeken, J. & Matthijs, G. Congenital disorders of glycosylation: a rapidly expanding disease family. Annu. Rev. Genom. Hum. Genet. 8, 261–278 (2007).
Peanne, R. et al. Congenital disorders of glycosylation (CDG): Quo vadis? Eur. J. Med. Genet. 30494–9 (2017).
Scott, K., Gadomski, T., Kozicz, T. & Morava, E. Congenital disorders of glycosylation: new defects and still counting. J. Inherit. Metab. Dis. 37, 609–617 (2014).
Ferreira, C. R. et al. Recognizable phenotypes in CDG. J. Inherit. Metab. Dis. 41, 541–553 (2018).
Schiff, M. et al. Clinical, laboratory and molecular findings and long-term follow-up data in 96 French patients with PMM2-CDG (phosphomannomutase 2-congenital disorder of glycosylation) and review of the literature. J. Med. Genet. 54, 843–851 (2017).
Chantret, I. et al. A deficiency in dolichyl-P-glucose: Glc1Man9GlcNAc2-PP-dolichyl alpha3-glucosyltransferase defines a new type of congenital disorders od glycosylation (CDG). J. Biol. Chem. 278, 9962–9971 (2003).
Charlwood, J. et al. A case of the carbohydrate-deficient glycoprotein syndrome type 1 (CDGS type 1) with normal phosphomannomutase activity. J. Inherit. Metab. Dis. 20, 817–826 (1997).
Eklund, E. A. et al. Congenital disorder of glycosylation (CDG)-Ih patient with a severe hepato-intestinal phenotype and evolving central nervous system pathology. J. Pediatr. 147, 847–850 (2005).
Schollen, E. et al. Clinical and molecular features of three patients with congenital disorders of glycosylation type Ih (CDG-Ih) (ALG8 deficiency). J. Med. Genet. 41, 550–556 (2004).
Vesela, K. et al. A new case of ALG8 deficiency (CDG Ih). J. Inherit. Metab. Dis 32, (Suppl 1), 259–264 (2009).
Stolting, T. et al. Novel ALG8 mutations expand the clinical spectrum of congenital disorder of glycosylation type Ih. Mol. Genet. Metab. 98, 305–309 (2009).
Bastaki, F. et al. Single-center experience of N-linked congenital disorders of glycosylation with a summary of molecularly characterized cases in Arabs. Ann. Hum. Genet. 82, 35–47 (2018).
Hock, M. et al. ALG8-CDG: novel patients and review of the literature. Orphanet J. Rare Dis. 10, 73 (2015).
Noensie, E. N. & Dietz, H. C. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 19, 434–439 (2001).
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Ng, P. C. & Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874 (2001).
Tavtigian, S. V., Pierotti, M. A. & Borresen-Dale, A. L. International Agency for Research on Cancer Workshop on ‘Expression array analyses in breast cancer taxonomy’. Breast Cancer Res. 8, 303 (2006).
Schwarz, J. M., Cooper, D. N., Schuelke, M. & Seelow, D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362 (2014).
Desmet, F. O. et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Lindahl, E., Azuara, C., Koehl, P. & Delarue, M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 34, W52–W56 (2006).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Frischmeyer, P. A. & Dietz, H. C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8, 1893–1900 (1999).
Hiller, M. et al. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 36, 1255–1257 (2004).
Hiller, M. & Platzer, M. Widespread and subtle: alternative splicing at short-distance tandem sites. Trends Genet 24, 246–255 (2008).
Dou, Y., Fox-Walsh, K. L., Baldi, P. F. & Hertel, K. J. Genomic splice-site analysis reveals frequent alternative splicing close to the dominant splice site. RNA 12, 2047–2056 (2006).
Ermakova, E. O., Nurtdinov, R. N. & Gelfand, M. S. Overlapping alternative donor splice sites in the human genome. J. Bioinform. Comput. Biol. 5, 991–1004 (2007).
Hamid, F. M. & Makeyev, E. V. Emerging functions of alternative splicing coupled with nonsense-mediated decay. Biochem. Soc. Trans. 42, 1168–1173 (2014).
Acknowledgments
This work was supported by a grant from ANR-15RAR3-0004-06 under the frame of E-RARE-3, the ERA-Net for Research on Rare Diseases.
Author information
Authors and Affiliations
Contributions
All authors have been involved in conception and design, or analysis and interpretation of data, and drafting the article or revising it critically for important intellectual content, and give their agreement to submission. S.V.-B., J.D., T.D., N.S., S.M., and I,C, are involved in analysis and interpretation of biochemistry and molecular data. M.S., E.S., A.D.H.O. d.B., P.d.L. collected clinical data. F.M. analyzed exome data for patient 2. A. C. performed 3D modeling data for normal and mutated protein of patient 1.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Vuillaumier-Barrot, S., Schiff, M., Mattioli, F. et al. Wide clinical spectrum in ALG8-CDG: clues from molecular findings suggest an explanation for a milder phenotype in the first-described patient. Pediatr Res 85, 384–389 (2019). https://doi.org/10.1038/s41390-018-0231-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-018-0231-5