Abstract
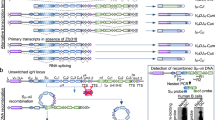
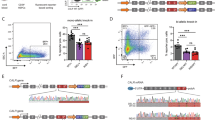
The error-prone V(D)J recombination process generates considerable amounts of nonproductive immunoglobulin (Ig) pre-mRNAs. We recently demonstrated that aberrant Ig chains lacking variable (V) domains can be produced after nonsense-associated altered splicing (NAS) events. Remarkably, the expression of these truncated Ig polypeptides heightens endoplasmic reticulum stress and shortens plasma cell (PC) lifespan. Many questions remain regarding the molecular mechanisms underlying this new truncated Ig exclusion (TIE-) checkpoint and its restriction to the ultimate stage of B-cell differentiation. To address these issues, we evaluated the extent of NAS of Ig pre-mRNAs using an Ig heavy chain (IgH) knock-in model that allows for uncoupling of V exon skipping from TIE-induced apoptosis. We found high levels of V exon skipping in PCs compared with B cells, and this skipping was correlated with a biallelic boost in IgH transcription during PC differentiation. Chromatin analysis further revealed that the skipped V exon turned into a pseudo-intron. Finally, we showed that hypertranscription of Ig genes facilitated V exon skipping upon passive administration of splice-switching antisense oligonucleotides (ASOs). Thus, V exon skipping is coupled to transcription and increases as PC differentiation proceeds, likely explaining the late occurrence of the TIE-checkpoint and opening new avenues for ASO-mediated strategies in PC disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Li, S. & Wilkinson, M. F. Nonsense surveillance in lymphocytes? Immunity 8, 135–141 (1998).
Deiss, T. C. et al. Immunogenetic factors driving formation of ultralong VH CDR3 in Bos taurus antibodies. Cell Mol. Immunol. https://doi.org/10.1038/cmi.2017.117 (2017).
Baumann, B., Potash, M. J. & Köhler, G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 4, 351–359 (1985).
Jäck, H. M., Berg, J. & Wabl, M. Translation affects immunoglobulin mRNA stability. Eur. J. Immunol. 19, 843–847 (1989).
Chemin, G. et al. Multiple RNA surveillance mechanisms cooperate to reduce the amount of nonfunctional Ig kappa transcripts. J. Immunol. 184, 5009–5017 (2010).
Tinguely, A. et al. Cross talk between immunoglobulin heavy-chain transcription and RNA surveillance during B cell development. Mol. Cell Biol. 32, 107–117 (2012).
Valentine, C. R. The association of nonsense codons with exon skipping. Mutat. Res. 411, 87–117 (1998).
Maquat, L. E. NASty effects on fibrillin pre-mRNA splicing: another case of ESE does it, but proposals for translation-dependent splice site choice live on. Genes Dev. 16, 1743–1753 (2002).
Chang, Y.-F., Chan, W.-K., Imam, J. S. & Wilkinson, M. F. Alternatively spliced T-cell receptor transcripts are up-regulated in response to disruption of either splicing elements or reading frame. J. Biol. Chem. 282, 29738–29747 (2007).
Srour, N. et al. A plasma cell differentiation quality control ablates B cell clones with biallelic Ig rearrangements and truncated Ig production. J. Exp. Med. 213, 109–122 (2016).
Delpy, L., Le Bert, M., Cogné, M. & Khamlichi, A. A. Germ-line transcription occurs on both the functional and the non-functional alleles of immunoglobulin constant heavy chain genes. Eur. J. Immunol. 33, 2108–2113 (2003).
Reynaud, S. et al. Interallelic class switch recombination contributes significantly to class switching in mouse B cells. J. Immunol. 174, 6176–6183 (2005).
Casola, S. et al. B cell receptor signal strength determines B cell fate. Nat. Immunol. 5, 317–327 (2004).
Lechouane, F. et al. B-cell receptor signal strength influences terminal differentiation. Eur. J. Immunol. 43, 619–628 (2013).
Wesemann, D. R. et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature 501, 112–115 (2013).
Aouinti, S. et al. IMGT/StatClonotype for pairwise evaluation and visualization of NGS IG and TR IMGT clonotype (AA) diversity or expression from IMGT/HighV-QUEST. Front. Immunol. 7, 339 (2016).
Pan, Q. et al. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 20, 153–158 (2006).
de la Mata, M. et al. RNA polymerase II elongation at the crossroads of transcription and alternative splicing. Genet. Res. Int. 2011, 309865 (2011).
Schor, I. E., Rascovan, N., Pelisch, F., Alló, M. & Kornblihtt, A. R. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl Acad. Sci. USA 106, 4325–4330 (2009).
Park, S.-K., Xiang, Y., Feng, X. & Garrard, W. T. Pronounced cohabitation of active immunoglobulin genes from three different chromosomes in transcription factories during maximal antibody synthesis. Genes Dev. 28, 1159–1164 (2014).
Kassambara, A. et al. GenomicScape: an easy-to-use web tool for gene expression data analysis. Application to investigate the molecular events in the differentiation of B cells into plasma cells. PLoS Comput. Biol. 11, e1004077 (2015).
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41, 376–381 (2009).
Meda, F., Folci, M., Baccarelli, A. & Selmi, C. The epigenetics of autoimmunity. Cell Mol. Immunol. 8, 226–236 (2011).
Luco, R. F. et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010).
Saint-André, V., Batsché, E., Rachez, C. & Muchardt, C. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat. Struct. Mol. Biol. 18, 337–344 (2011).
Zhou, H.-L., Luo, G., Wise, J. A. & Lou, H. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 42, 701–713 (2014).
Benchaouir, R., Robin, V. & Goyenvalle, A. Gene and splicing therapies for neuromuscular diseases. Front. Biosci. Landmark Ed. 20, 1190–1233 (2015).
Groen, E. J. N., Talbot, K., Gillingwater, T. H. Advances in therapy for spinal muscular atrophy: promises and challenges. Nat. Rev. Neurol. https://doi.org/10.1038/nrneurol.2018.4 (2018).
Stein, C. A. & Castanotto, D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. J. Am. Soc. Gene Ther. 25, 1069–1075 (2017).
Mostoslavsky, R., Alt, F. W. & Rajewsky, K. The lingering enigma of the allelic exclusion mechanism. Cell 118, 539–544 (2004).
Blencowe, B. J. Alternative splicing: new insights from global analyses. Cell 126, 37–47 (2006).
Johnson, K., Angelin-Duclos, C., Park, S. & Calame, K. L. Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell Biol. 23, 2438–2450 (2003).
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Aartsma-Rus, A. & van Ommen, G.-J. B. Progress in therapeutic antisense applications for neuromuscular disorders. Eur. J. Hum. Genet. 18, 146–153 (2010).
Daly, J., Licence, S., Nanou, A., Morgan, G. & Mårtensson, I.-L. Transcription of productive and nonproductive VDJ-recombined alleles after IgH allelic exclusion. EMBO J. 26, 4273–4282 (2007).
Delpy, L., Sirac, C., Le Morvan, C. & Cogné, M. Transcription-dependent somatic hypermutation occurs at similar levels on functional and nonfunctional rearranged IgH alleles. J. Immunol. 173, 1842–1848 (2004).
Holwerda, S. J. B. et al. Allelic exclusion of the immunoglobulin heavy chain locus is independent of its nuclear localization in mature B cells. Nucleic Acids Res. 41, 6905–6916 (2013).
Nogués, G. et al. Control of alternative pre-mRNA splicing by RNA Pol II elongation: faster is not always better. IUBMB Life 55, 235–241 (2003).
Pandit, S., Wang, D. & Fu, X.-D. Functional integration of transcriptional and RNA processing machineries. Curr. Opin. Cell Biol. 20, 260–265 (2008).
Shukla, S. & Oberdoerffer, S. Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim. Biophys. Acta 1819, 673–683 (2012).
Pinaud, E. et al. Localization of the 3’ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity 15, 187–199 (2001).
Sirac, C. et al. Role of the monoclonal kappa chain V domain and reversibility of renal damage in a transgenic model of acquired Fanconi syndrome. Blood 108, 536–543 (2006).
Rouaud, P. et al. The IgH 3’ regulatory region controls somatic hypermutation in germinal center B cells. J. Exp. Med. 210, 1501–1507 (2013).
Green, M. R. et al. Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc. Natl Acad. Sci. USA 108, 2873–2878 (2011).
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001).
Li, S. et al. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat. Commun. 4, 2333 (2013).
Boice, M. et al. Loss of the HVEM tumor suppressor in lymphoma and restoration by modified CAR-T cells. Cell 167, 405–418.e13 (2016).
Alamyar, E., Duroux, P., Lefranc, M.-P. & Giudicelli, V. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 882, 569–604 (2012).
Bischof, J. & Ibrahim, S. M. bcRep: R package for comprehensive analysis of B cell receptor repertoire data. PLoS ONE 11, e0161569 (2016).
Acknowledgements
We thank the staff of our animal facility as well as C. Carrion for technical assistance with microscopy and cell cytometry. We also thank K. Rajewsky (The Max Delbrück Center for Molecular Medicine, Berlin, Germany) and S. Casola (Institute of Molecular Oncology Foundation, Milano, Italy) for providing DH-LMP2A mice. We are grateful to N. Diallon (MRC Clinical Sciences Centre, London, UK) for providing the pγSat plasmid and M. Alizadeh (UMR S 917, Rennes, France) for repertoire sequencing. This work was supported by grants from Fondation ARC (PJA 20161204724/PGA120150202338), INCa (PLBIO15-256), ANR (2017-CE15-0024-01), Ligue Contre le Cancer (comités Corrèze, Haute-Vienne), Fondation Française pour la Recherche contre le Myélome et les Gammapathies monoclonales (FFRMG) and Comité d’Organisation de la Recherche sur le Cancer du Limousin (CORC). M.O.A. and N.S. were funded by Société Française d’Hématologie (SFH) and Région Limousin, respectively.
Author information
Authors and Affiliations
Contributions
M.O.A. and N.S. designed and performed experiments, analyzed data and wrote the paper. J.-M.L. and A.M. contributed data to Fig. 6. O.M., S.L.N., and E.P. contributed 3D DNA-FISH data in Fig. 3. J.M. performed gene expression analysis in Fig. 4. M.V.A., J.S., C.S., and M.C. helped with the experiments and interpretation of the data. L.D. conceived the project, designed experiments, analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Ashi, M.O., Srour, N., Lambert, JM. et al. Physiological and druggable skipping of immunoglobulin variable exons in plasma cells. Cell Mol Immunol 16, 810–819 (2019). https://doi.org/10.1038/s41423-018-0160-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-018-0160-6
Keywords
This article is cited by
-
Mediator contributes to IgH locus VDJ rearrangements by promoting usage of most distal V segments
Cellular & Molecular Immunology (2020)